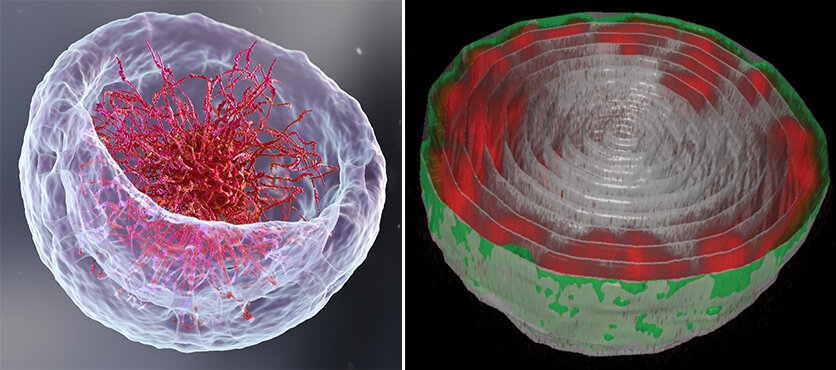
Left: An illustration of the nucleus that represents the classical theory about DNA organization. Right: Turning the bowl of ramen on its headMicroscopic view of the nucleus in a fruit fly larvae's muscle cell. The DNA chains (red) are attached the the nuclear lamina, the inner layer of nuclear membrane. Credit: Weizmann Institute of Science
You'll feel hungry if you open a biology book and look at the pictures of how DNA is structured in cells' nucleus. The DNA chains would be like a bowl full of ramen, with long strings suspended in liquid. This image needs to be rethought, according two new experiments and one theoretical study that were the result of collaboration between Prof. Talila Volks of the Molecular Genetics Department and Professor. Sam Safran of Chemical and Biological Physics Department of the Weizmann Institute of Science. It is important to clarify this image, as the spatial arrangement of DNA in the nucleus can influence the expression of the genes and therefore the protein found in cells.
Volk began this story when she was researching how mechanical forces affect cell nuclei in muscle cells and discovered that muscle contractions have an immediate effect upon gene expression patterns. She says, "We couldn’t explore this further because existing imaging methods depended on imaging chemically preserved cells, so these failed to capture the actual cell nuclei of a working muscle."
Dr. Dana Lorber is a research associate in Volk’s group and designed a device to allow for the study of muscle nuclei in live fruit flies larvae. The tiny, transparent larva is held in a groove that allows them to relax and contract their muscles. However, the device restricts its movements so that a fluorescence microscope can scan it. Researchers obtained images of the complex of DNA and proteins within the device. They were surrounded by the membrane that contains the muscle nuclei.
Lorber and Daria Amiad Pavlov, a postdoctoral fellow from Volk's group, expected a bowl of ramen. Instead of filling the entire nucleus with chromatin molecules, the "noodles", or long chromatin molecules were attached to the nucleus' inner walls as a thin layer. The chromatin was able to separate itself from the bulk liquid within the nucleus, a process known as "phase separation". It found its place near the edges of the nucleus, while the majority of the fluid medium remained in the middle. They realized they were close to solving a fundamental biological question: how is chromatin and DNA organized in the nucleus of a living organism? Lorber states that the unexpected findings forced them to check for errors and ensure that the organization was universal.
3D chromatin simulations show that the physical interaction between the nuclear lamina and chromatin is crucial for the organization of chromatin in the nucleus. These interactions can weaken (left-to-right), as is the case with many diseases, including muscle dystrophies and neurological disorders. The chromatin moves from the nucleus' periphery to its center. Credit: Weizmann Institute of Science
They came to the conclusion that there had been no mistake after collaborating with Safran's research group. Safran and Gaurav Bajpai, a postdoctoral fellow, created a theoretical model of chromatin organization in nucleus. It included physical factors such as the relative forces and attraction between chromatin (and its liquid environment) and chromatin (and the nuclear membrane). Based on the amount of liquid in the nucleus, the model predicted that the chromatin would separate from the liquid phase. The phase separated chromatin could then be organized along the nuclear membrane, just as Volk's research team found.
These groups also explained why chromatin filled the cell nuclei in previous research by scientists. Scientists can flatten cells by placing them on a glass slide to examine them under a microscope. Safran explains that this may alter some forces governing chromatin arrangement, and reduce the distance between its base and the upper part.
Lorber and Amiad Pavlov teamed up with Francesco Roncato, a member of Prof. Ronen Alon’s Immunology Department, to examine live human white blood cell samples in order to ensure that these findings weren't limited only to fruit fly muscle cells. The chromatin in this case was also organized in a layer that lined the inner nuclear wall. Amiad Pavlov says, "This demonstrated that the phenomenon we had found was likely to be widespread and that this chromatin organization has probably been conserved through evolution."
This study opens new research avenues into DNA's organization within cells and, in turn, the physical forces that affect the nucleus, chromatin, and gene expression. One possibility is to determine if there's a difference in DNA organization between healthy and sick. This difference could be used to diagnose cancer cells, such as by using it as a new method of detecting them. Exploring DNA organization in embryonic development may reveal whether mechanical forces influence the differentiation of cells into different fates. It is known that the stiffness of the cell surface can affect the expression of genes. This may be due to the force and pull of the surface on the nuclear membrane, and its impact on DNA organization within a nucleus. This interplay can be used to control gene expression in cells that are used for engineering tissues with desirable properties.
These studies were published in Science Advances as well as eLife.
Explore more Discovery discovers that a cellular building blocks acts as a gel and not as liquid as previously thought
More information: Daria Amiad-Pavlov et al, Live imaging of chromatin distribution reveals novel principles of nuclear architecture and chromatin compartmentalization, Science Advances (2021). Daria Amiad-Pavlov et al, Live imaging of chromatin distribution reveals novel principles of nuclear architecture and chromatin compartmentalization,(2021). DOI: 10.1126/sciadv.abf6251 Gaurav Bajpai et al, Mesoscale phase separation of chromatin in the nucleus, eLife (2021). DOI: 10.7554/eLife.63976 Journal Information: Science Advances
