It's not enough to know the structure of a biological system. Knowing how the system moves is important.
Rice University researchers have created a model of a key mechanism.
Combining structural experiments and computer simulations, bioscientist Yang Gao, theoretical physicist Peter Wolynes, graduate student ShiKai Jin, and their colleagues have uncovered details about how helicases, a family of ringlike motor proteins, wrangle DNA during replication. New targets for disease fighting drugs could be revealed by their work.
The synergy between the experiments and large-scale simulations they describe could become a paradigm for modeling of the mechanisms of many complex biological systems.
"These are dynamic processes that can't be captured with experimental methods alone," said Gao, an assistant professor of biosciences. It's important to show the mechanisms of the helicases, because they're essential for the replication of genes.
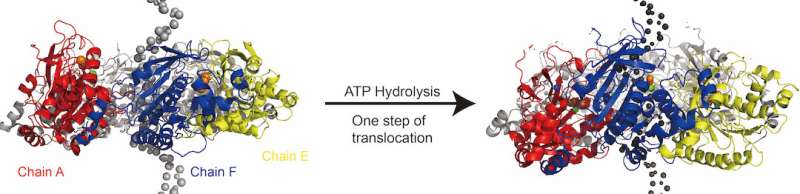
A washer-like ring that separates the parents' double strands of DNA from the daughter's single strands is what a hydrameric helicase looks like. Researchers haven't been able to pin down how the helicase moves as it opens.
It was known that the stepping movement was driven by the NTPs. Researchers didn't know that the key wasATP hydrolysis. The Rice team found that the binding of the helicase subunits was tightly bound, but that the energy barrier for disassociation was greatly lowered.
The researchers noted that because the helicase is so large, only a few attempts have been made to transfer it from one strand to another. The opportunity to study the process from start to finish was provided by the Rice hybrid.
Several previously unknown intermediate states were revealed in the simulations. Each step of the process can carry more than 12 nucleotides.
The team focused on the T7 bacteriophage as a model system to look for the mechanism. The researchers combined two force fields to create a simulation of the helicase. The forces that control how atoms and Molecules move are described in force fields. A paper on the novel combination of force fields was written by Rice.
Machine learning uses a subset of the atoms in a system but still provides accurate results while cutting computation time.
We could make our simulations very fast by combining the models. The combined software is 30 times faster than previous versions.
T7 is half the size of a human cell. He said that in human systems, there are six different polypeptide chains, but in T7, it's the same one that makes six copies.
"Because our new form of open3SPN2 deals with a single strand of DNA, it allows us to analyze processes where the normally double-stranded DNA opens up as it does in the presence of the helicase," he said. The single-stranded DNA force field is novel but this was just a background in the project where we can look at the process in great detail.
The structures we have for these complexes are very precise. There's still a lot we don't know about how they do that.
Computational models can make a big contribution and they will be adapted to other large systems to examine important questions.
More information: Shikai Jin et al, Computationally exploring the mechanism of bacteriophage T7 gp4 helicase translocating along ssDNA, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2202239119