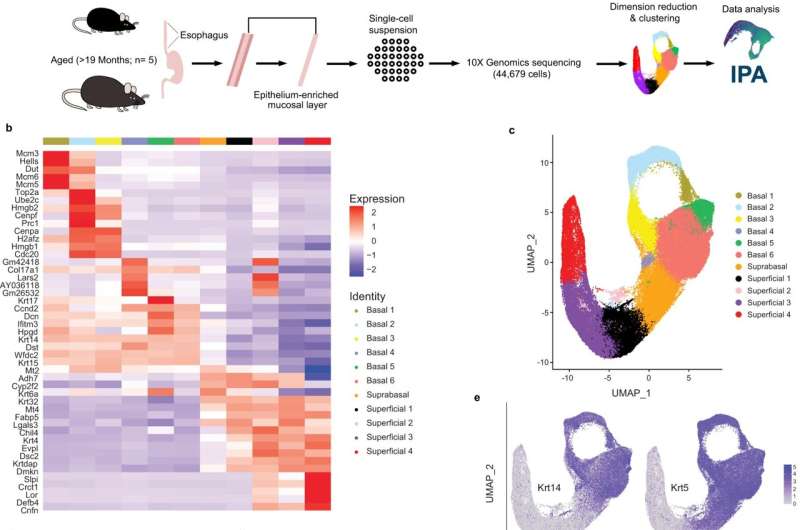
In a study published today in the journal Nature Communications, researchers at the Lewis Katz School of Medicine at Temple University defined subsets of cells found in the esophagus of mice, information that could potentially help clinicians diagnose or treat certain types of cancer.
The tissue in the esophagus is comprised of a basal layer that eventually gives rise to several layers of differentiated cells. Researchers wanted to find out if the cells were the same or if there were differences in their expression.
This work lays the foundation to address questions about how these cells relate to the development of cancer, according to Kelly Whelan, PhD, senior author on the study and assistant professor of cancer and cellular biology at the Lewis.
The next question is if we give a mouse a carcinogen, does one subset expand or is another subset deplete? We would want to know whether cell types that change in the context of cancer are contributing to the cancer process and whether we can target them to help treat or prevent cancer.
To address these questions, the researchers used single cell gene-expression profiling, a tool that identifies all the genes in a cell or tissue. Individual cell types can be identified with the help of these profiles.
She said that they took more than 40,000 cells from the mouse and used their individual genes to group them.
When the researchers started the study, they expected to find three subsets of cells, but instead they found six, as well as four that were differentiated. One cell type was found to have characteristics of both differentiated and basal cells and was defined as an intermediate cell type.
We are trying to figure out what the cells do in the normal context and in the context of cancer by isolating these individual cell types. The study is a fundamental step towards that.
More information: Mohammad Faujul Kabir et al, Single cell transcriptomic analysis reveals cellular diversity of murine esophageal epithelium, Nature Communications (2022). DOI: 10.1038/s41467-022-29747-x Journal information: Nature Communications Citation: Researchers reveal cellular diversity of esophageal tissue (2022, April 22) retrieved 22 April 2022 from https://phys.org/news/2022-04-reveal-cellular-diversity-esophageal-tissue.html This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.