Ingrid Fadelli is a writer for Phys.org.
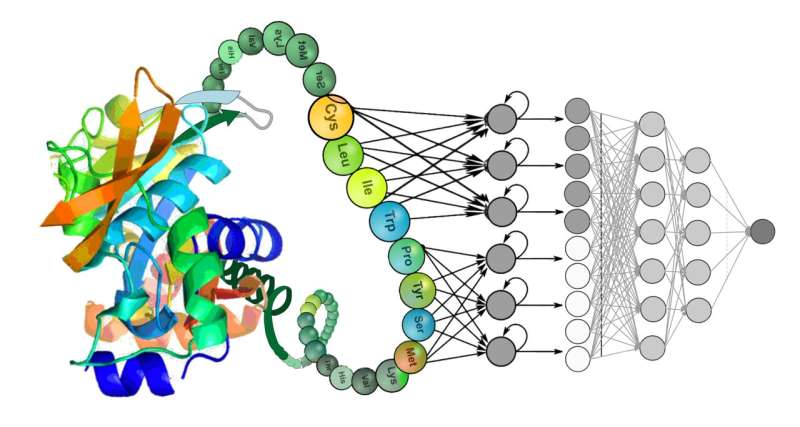
BERT, T5 and GPT are examples of deep learning language models that can be used to analyze speech and texts. They have been used in the fields of biomedicine and biotechnology to study genetic codes.
Genetics researchers and bioinformaticians have been trying to understand the biological roles of genes for decades. They need to analyze large and complex biological data to do this.
Researchers at Hacettepe University, Middle East Technical University and Karadeniz Technical University, Turkey, have recently carried out a study evaluating the potential of deep learning based language models for studying and predicting the functional properties of proteins. The advantages and disadvantages of different state-of-the-art approaches are summarized in their paper.
The sequence of a genes can be thought of as a sentence with a specific meaning in natural language, if the data is modeled as a language.
The ability of different modeling approaches to extract hidden patterns of the functional properties of the proteins was assessed in the paper. Their evaluations included all of the most well-known natural language modeling architectures, each of which can contain hundreds of millions or billions of parameters.
Self-supervised pre-training of these models requires huge resources.
The team had to build large and reliable testing datasets, each with a different difficulty level, in order to assess the models and compare their performances. They created four benchmark datasets that allowed them to investigate semantic similarities, functional definitions, and drug target families. All of these are important biological mechanisms that are linked to the occurrence and progression of different types of cancer.
The most notable finding was that the models were able to learn the functional properties of the proteins using the only input, the amino acid sequence, which is quite a difficult problem.
In the future, the models evaluated by this team of researchers could help to enhance precision medicine interventions, for instance analyzing the molecular make-up of patients resulting from genomic variations to develop personalized treatments. The results gathered by Do11F;an and his colleagues highlight the huge potential of deep learning, but existing methods need to be improved before they can be used in real-life clinical decision-making systems.
We are working on a new system to better represent proteins. Our ultimate goal is to get a universal representation that can be used in any modeling task.
More information: Serbulent Unsal et al, Learning functional properties of proteins with language models, Nature Machine Intelligence (2022). DOI: 10.1038/s42256-022-00457-9 Journal information: Nature Machine IntelligenceThe Science X Network will be launched in 2022.
Citation: Study evaluates deep learning models that decode the functional properties of proteins (2022, April 18) retrieved 18 April 2022 from https://phys.org/news/2022-04-deep-decode-functional-properties-proteins.html This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.