Max Helf was a research associate in the lab of Frank Schroeder, and he saw his labmates struggle when analyzing data. Metaboseek is a free, open-source app that is essential to the lab's work.
One of the most successful model systems for human biology, the roundworm Caenorhabditis elegans, is being studied by the Schroeder lab to discover new metabolites that govern evolutionarily conserved signaling pathways and could be useful as leads for the development of new pharmaceuticals or agrochemicals. The researchers compare the metabolites between two different worm populations.
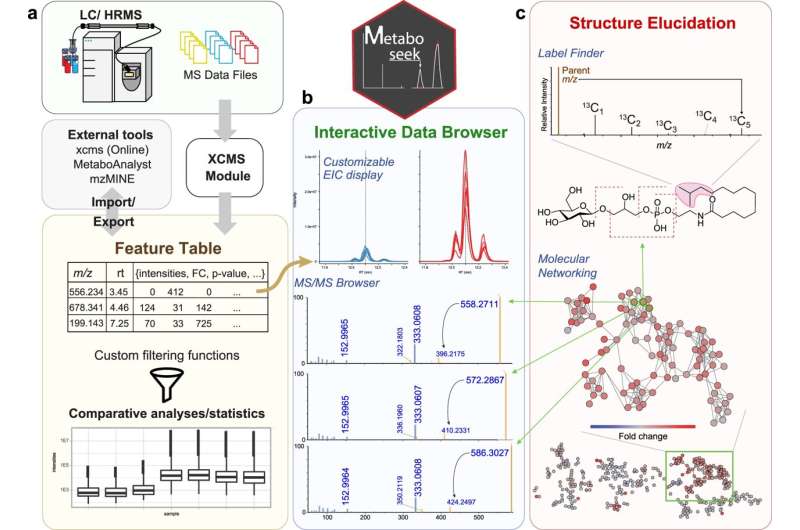
Computational approaches are needed to analyze samples that have more than 100,000 compounds.
The team had been relying on software packages that did not allow them to easily modify analysis parameters. The lack of a graphical user interface made it difficult for his colleagues to visually inspect mounds of data.
Helf thought there had to be an easier way to solve the problem.
After addressing the problems my labmates were already facing, I talked to them about what else held them back.
The result was Metaboseek, an app with a graphical interface that incorporates multiple data analysis tools that non-coding researchers wouldn't have. The app streamlines the analysis of comparative metabolomics data by helping the researcher determine which data features are real and letting them dig deeper into those features within the same tool.
"Max did this without me even requesting it." We started using it, and now our lab and many partners couldn't exist without it.
The proof-of-concept for Metaboseek was provided in a study published in Nature Communications on February 10.
The team used Metaboseek to discover that roundworms lacking a key gene in the α-oxidation pathway accumulated hundreds of previously unreported metabolites. The findings are important because they show that oxidation is a basic pathway in worms that is conserved in humans.
The study was a collaboration between the two postdocs and was done byBennett Fox, who was a professor in Cornell University's Department of Chemistry and Chemical Biology.
There are a few reasons why there aren't a lot of good analytic tools for comparing metabolomics data. There isn't enough time to develop software tools and database infrastructure because comparative metabolomics is a relatively young field.
Over the last decade, the advent of affordable, ultra-high-resolution mass spectrometers for collecting metabolomics data has increased by perhaps more than tenfold the amount of data one sample can generate, creating an even greater need for sophisticated tools that can keep up with.
Metaboseek has an array of features for analyzing various types of data to aid compound identification, structure determination, assignment of metabolites to families based on structural similarities, and more.
More information: Maximilian J. Helf et al, Comparative metabolomics with Metaboseek reveals functions of a conserved fat metabolism pathway in C. elegans, Nature Communications (2022). DOI: 10.1038/s41467-022-28391-9 Journal information: Nature Communications Citation: New software to help discover valuable compounds (2022, March 29) retrieved 29 March 2022 from https://phys.org/news/2022-03-software-valuable-compounds.html This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.