MIT chemists have found the structure of a molecule that can be pumped out of a cell. This one is found in E. coli and is believed to helpbacteria become resistant to antibiotics.
The researchers were able to determine how the structure of thisProtein changes as a drug-like molecule moves through it. According to an MIT professor of chemistry, knowledge of this structure may make it possible to design drugs that could block these transport proteins and help resensitize drug-resistantbacteria to existing antibiotics.
Hong, who is the senior author of the paper, says that knowing the structure of the drug-binding pocket could be used to design competitors that would block the binding site.
Alexander Shcherbakov is a graduate student at MIT. The research team includes a graduate student from MIT and a professor from the University of Wisconsin at Madison.
Drug-resistance transporters.
One of the ways that antibiotics can be evaded is by pumping drugs out of their cells. The group at the University of Wisconsin has been studying EmrE for several years, and it can transport many different toxic compounds.
EmrE is a member of the small multidrug resistance (SMR) transporters. EmrE is not involved in resistance to antibiotics, but other members of the family have been found in drug-resistant forms.
The SMR transporters have high sequence conserved across key regions. EmrE is by far the best-studied member of the family and it is an ideal model system to investigate the structure that supports SMR activity.
A few years ago, Hong's lab developed a technique that allows researchers to use NMR to measure the distances between fluorine probes and hydrogen atoms. It's possible to determine the structure of aProtein as it binding to a molecule with fluorine
After Hong gave a talk about the new technique at a conference, Henzler-Wildman suggested that they team up to study EmrE. Her lab has spent many years studying how EmrE transports a drug-like molecule. F 4 -TPP + is a molecule with four fluorine atoms attached to it, one at each corner.
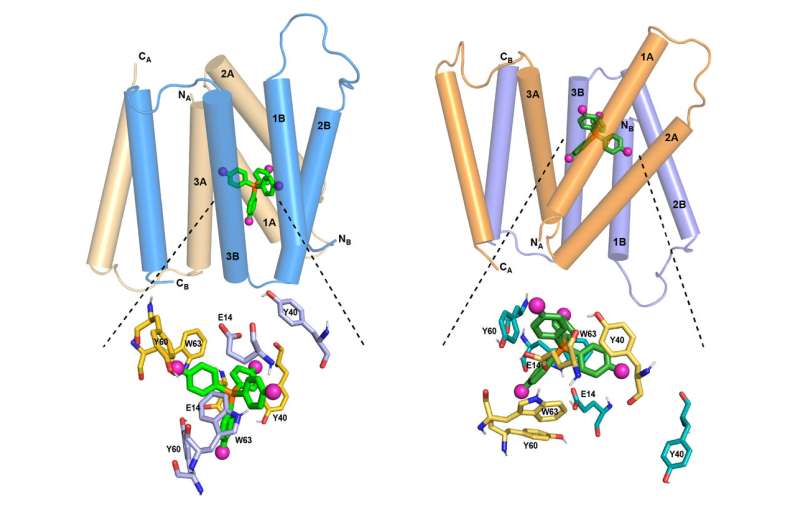
The researchers set out to determine an atomic-resolution structure of EmrE using the new NMR technique. Each EmrE molecule has four transmembrane helices that are 888-609- 888-609- 888-609- 888-609- Two EmrE molecule assemble into a dimer, so that eight transmembrane helices form inner walls that interact with the ligand as it moves through the channel. The overall helices have been revealed, but not the side chains that extend into the channel interior, which are like arms.
EmrE transports toxic molecule from the inside of a cell to the outside, which is acidic. The structure of EmrE is affected by the change in pH. Hong and Henzler-Wildman discovered the structure of theProtein as it binding to F 4 -TPP + in an acidic environment. In the new study, they were able to determine how the structure of the protein changes as the pH changes.
A complete structure.
The researchers found that the four helices that make up the channel are close to each other, making it easy for the ligand to enter. The helices tilt as the pH drops so that the channel is more open to the outside of the cell. This helps to get the ligand out of the channel. Several rings found in the side chains help to guide the ligand out of the channel.
The acidic end of the channel is more welcoming to protons, which help it to open further, allowing the ligand to exit more easily.
Hong says the paper really completes the story. To figure out how a transporter can open to both sides of the cell, you need two.
Hong and her colleagues are going to study how other compounds travel through the EmrE channel.
More information: High-pH Structure of EmrE Reveals the Mechanism of Proton- Coupled Substrate Transport, Nature Communications (2022). DOI: 10.1038/s41467-022-28556-6 Journal information: Nature Communications Citation: Protein structure offers clues to drug-resistance mechanism (2022, February 18) retrieved 18 February 2022 from https://phys.org/news/2022-02-protein-clues-drug-resistance-mechanism.html This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.