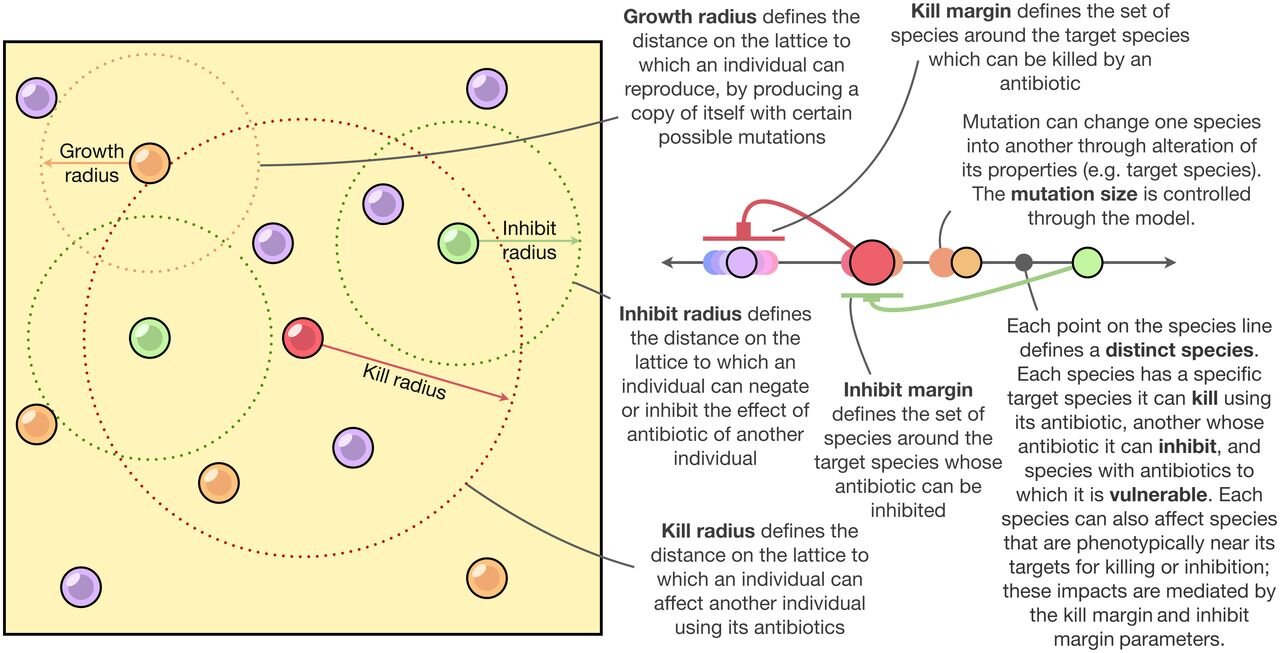
A conceptual representation of the model. Credit is given to 10.1073/pnas.
The University of Maryland biologists created the first mathematical simulations of the complex interactions and rapid evolution ofbacteria and reflected the tremendous species diversity seen in real life.
Their work was published in the Proceedings of the National Academy of Sciences on January 4, 2022.
It's like trying to understand wildlife in a world where elephants with gray skin and elephants with feathers tussle for space at the watering hole next to birds with trunks, zebras with scales and lions that evolved a taste for grass. New animals may appear with more than one trait the next day.
Thousands of species can exist side by side in a fluid state of evolution. Individualbacteria snatch up bits of DNA from their neighbors and quickly acquire new characteristics. The community is in a constant state of war because of complex alliances. Some species kill each other with toxins, while others shield each other from attackers.
Traditional theoretical models have not been effective for explaining how the complex interactions between species and each other work.
Microbial communities are important to human health and life on Earth. Microbes help regulate the immune system and cause and prevent disease. A balanced response to antibiotics is dependent on species diversity in the community.
Competition between species should lead to the rise of a few dominant species, according to conventional rules of biology. The rules don't reflect reality. They only account for interactions between two species at a time and don't consider the rapid speed of evolution seen in microbes.
The lack of an appropriately sophisticated theoretical framework that accounts for the complex rules driving community dynamics has hindered attempts to develop predictions that scientists can test with real-world experiments.
A petri dish or kitchen counter are examples of where a simulation community is spreading. Different colors represent different species, with more closely related species appearing more similar in color. The first to show the evolution of a community are the UMD models, which are different from traditional mathematical models. Credit: Anshuman.
"There has been very little theory that includes complex interactions to tell the experimentalists what is possible, what to look for, or how to expect a community to behave under certain conditions," he said. It's difficult to do experiments with many species, all of which are interacting with the community. How do you keep up with the rapid changes?
The computer simulations that were developed incorporated the unique complexities of this problem. After running over 10 million simulations, the team identified three key factors that influence species diversity, pointing for the first time to aspects of community dynamics that experimentalists can focus on.
There are higher-order interactions that are regulated by one or more additional species. species A creates a toxin that kills species B, but species C counteracts that toxin if B is still alive.
A "continuous trait space" is created by horizontal gene transfer. A community with a continuum of shared traits is caused by genetic mixing.
There are high rates of variation among species.
The researchers focused on these three factors and created a simulation to model the growth and evolution of the community. Their models accounted for unprecedented diversity in communities with a lot of different species. Traditional mathematical models that tried to incorporate complex interactions between species were usually limited to five or fewer distinct species.
The time it took for a community to reach equilibrium was the best predictor of species diversity, according to the models used by the group. A few species dominated when a community was stable.
Diversity flourished somewhere in between. The amount of diversity in a community was dictated by the amount of variation in the bacterium in the community.
With this framework, experimentalists should be better prepared to address important questions, like how to develop antibiotics that maintain diversity in the gut microbiome or how to prevent antibiotic resistant strains ofbacteria from dominating in an infection.
The research paper was published in PNAS on January 4, 2022.
Anshuman Swain et al., Higher-order effects, continuous species interactions, and trait evolution shape the dynamics of the Microbial Spatial Dynamics, Proceedings of the National Academy of Sciences. There is a book titled "10.1073/pnas.2020956119."
The National Academy of Sciences has a journal.
Rethinking the wild world of species diversity in microbes was retrieved fromphys.org on January 5, 2022.
The document is copyrighted. Any fair dealing for the purpose of private study or research cannot be reproduced without written permission. The content is not intended to be used for anything other than information purposes.
