UC Berkeley scientists had to develop two new methods to successfully edit genes within multiple members of a microbial community. The DART system is barcoded and compatible with the ET-Seq so that scientists can insert, track and assess the efficiency and specificity of their work. The lab is located at UC Berkeley.
In the past, the editing of the genomes of one type of cell at a time has been done with the help of the CRISPR enzymes.
A group at the University of California, Berkeley has found a way to add or modify genes within a community of many different species at the same time.
This technology can be used to track edited microbes in a natural community, such as in the gut or the roots of a plant, while still being only applied to lab settings. As scientists talk about how to change the population of microbes in order to fix problems, such tracking becomes necessary.
Without a way to track the genes, they could end up anywhere.
Benjamin Rubin said that breaking and changing DNA within isolated organisms has been essential to understanding what that DNA does. This work helps bring that fundamental approach to the community of microbes, which are much more representative of how they live and function in nature.
While the ability to "shotgun" edit many types of cells at once could be useful in current industry-scale systems, the more immediate application may be as a tool in understanding the structure of complex communities ofbacteria.
"Eventually, we may be able to eliminate genes that cause sickness in your gutbacteria or make plants more efficient by engineering their microbial partners," said Brady Cress. This approach will give us a better understanding of how the community works.
Rubin and Cress are both in the lab of CRISPR-Cas9 inventor, and Spencer Diamond is a project scientist in the Innovative Genomics Institute.
From censusing to editing.
Diamond works in the laboratory of a pioneer in the field of metagenomics, who pioneered the field of community sequencing, and who has never before done anything like this.
The past 15 years have seen the advancement of metagenomics. Diamond assembled 10,000 individual genomes from soil samples collected from a grassland meadow in Northern California.
He compares this to taking a population census, which gives unparalleled information about which microbes are present in which proportions, and which functions those microbes could perform within the community. It allows you to infer complicated interactions among organisms and how they may work together to achieve important benefits. New methods are needed to test these functions and interactions at a community level.
He said that the idea of metabolic handoffs is that no individual microbe is performing a huge string of metabolic functions, but for the most part, each individual organisms is doing a single step of a process. How do we prove this? How do we get to a point where we can actually see what's going on, instead of just watching the birds? This was the beginning of community editing.
The research team was led by Banfield, UC Berkeley professor of earth and planetary science and of environmental science, policy and management, as well as the Howard Hughes Medical Institute investigator and co-winner of the 2020 Nobel Prize in Chemistry, and they were joined by other researchers.
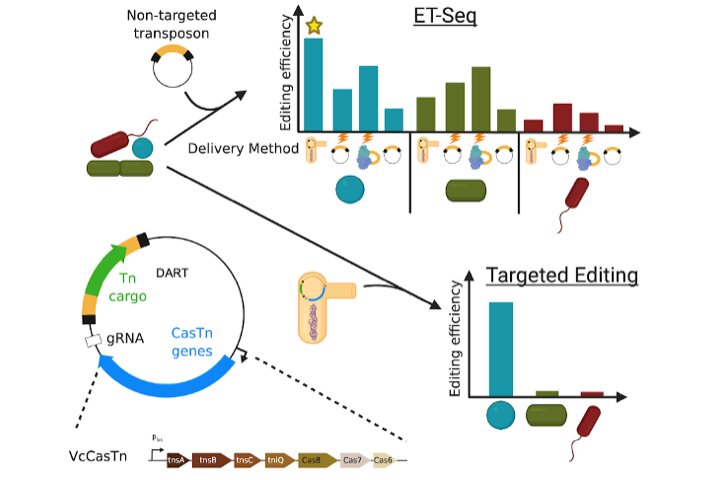
The approach developed by the team is to determine which microbes in the community are vulnerable to gene editing. Rubin and Diamond developed a screening technique that uses a transposon, or jumping gene, to find out if a certain type of bacterium is present. They were able to identify which species of microbes was able to incorporate the transposon by using the community DNA before and after introducing the transposon. The approach was based on techniques developed at Lawrence Berkeley National Laboratory. In one experiment, they successfully inserted the same transposon into five of the different microbes using different methods.
A bar-coded transposon is inserted into a specific DNA sequence through a targeted delivery system called All-in-oneRNA-guided CRISPR-Cas Transposase (DART).
The researchers cultured a stool sample from an infant and used it to create a community of 14 different types of microorganisms. They were able to modify individual strains within the community.
The researchers hope to use the technique to understand artificial, simple communities, such as a plant and its associated microbiome, in a closed box. They can track the effect on their bar-coded microbes by manipulating community genes within this closed system. The Department of Energy funded these experiments as part of a 10-year program called m-CAFEs, for Microbial Community Analysis and Functional Evaluation in Soils, which seeks to understand the response of a simple grass microbiome to external changes. The m-CAFEs project includes Banfield, Doudna, and Deutschbauer.
The other co-authors of the paper are Alexander and Christine He.
Benjamin E. Rubin and his team have published a paper on genome editing in complex bacterial communities. There is a DOI titled " 10.1038/s41564-021-01014-7".
The journal contains information about Nature Microbiology.
There is a news story on the topic of CRISPRing the microbiome that was retrieved fromphys.org on December 6th, 2011.
The document is copyrighted. Any fair dealing for the purpose of private study or research cannot be reproduced without written permission. The content is not intended to be used for anything other than information purposes.
