The University of Duesseldorf is headed by Arne Claussen.
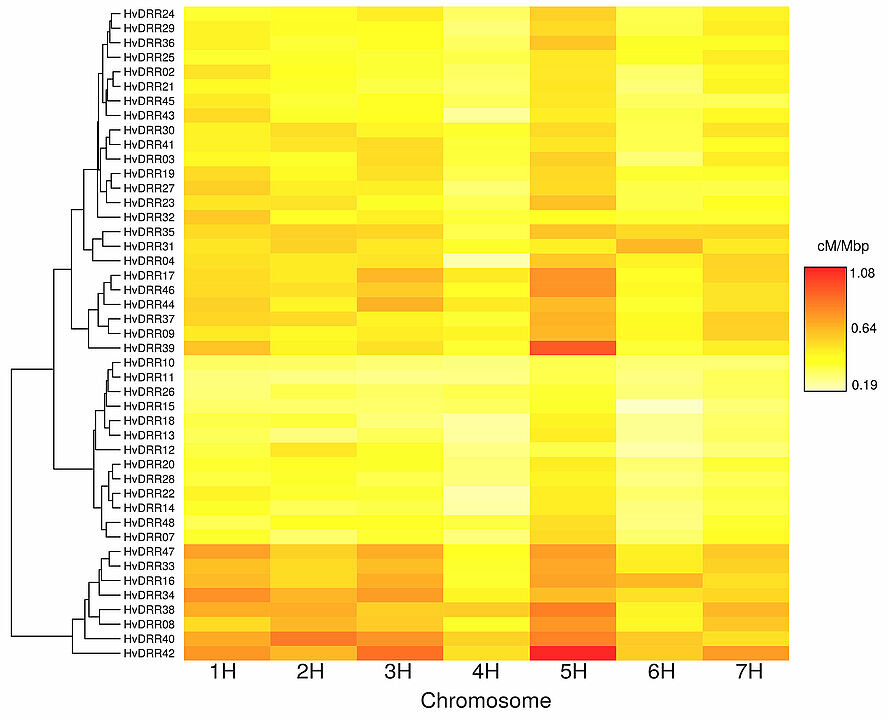
There are differences in the rate of recombination between the 45 spring barley populations, with red representing a high rate and white a low rate. Credit: HHU
It's important to create as many combinations as possible of genetic variations within a short time to find the most suitable candidates between plants with many different characteristics. A method for using natural variations to identify highly recombinogenic individuals has been developed by the working group of Prof. Dr. Benjamin Stich. The method was tested in a large-scale barley experiment.
The genes of the parents are rearranged during the sexual reproduction of organisms. The genome of the offspring's is a mosaic of its parents' genes. A new set of individual characteristics in the offspring are created by this genetic combination.
It's important to have as many genetic combination options as possible to breed new plant varieties. The capacity to generate variation in the progeny is increased by the fact that some genotypes rearrange their genome at more locations than others. Highly recombinogenic genotypes with this property are what they are called.
It is possible to insert a characteristic into a plant. A team of biologists from the HHU Institute of Quantitative Genetics and Genomics of Plants pursued a different approach. They developed a method to identify the highly recombinogenic genotypes in natural populations.
The idea of selecting plants that are naturally recombinogenic has been around for 70 years. It wasn't possible in practice because of the excessive experimental effort needed to find a few highly recombinogenic ones.
The researchers in Dsseldorf worked with 45 populations of spring barley. They looked at 50,000 markers in the offspring of those members to see how the parents' genomes were mixed. Their main finding was that the degree of mixing can vary by a factor of 2.5.
The effect of an individual parent can be found in 90 percent of the variation in offspring's recombination. In the case of a highly recombined population, it is clear that one or both of the parents is recombinogenic. It is possible to breed plants with increased recombination with this knowledge.
The researchers used a computer simulation to show that the differences in the recombination rate can be predicted with an accuracy of 80%. The method used in Dsseldorf makes it possible to design breeding selection programs much more efficiently and thus to accelerate the development of plant and animal varieties.
Prof. Stich said that to secure our food supply in the face of climate change, we need new variations of known food plants that are better equipped to deal with new environmental conditions and can produce a higher yield. The scientific foundations for this are in the CEPLAS. With our new method, we can now breed highly recombinogenic genotypes systematically to then produce considerably more variant in a crossing experiment to then select the most promising plants from that group.
The Genomic prediction of the recombination rate variation in barley is a route to highly recombinogenic genotypes. The DOI is 10.1111/pbi.13746.
There is information in the Plant Biotechnology Journal.
It was provided by the University of Duesseldorf.
There is a news story about highly-recombinant plants that was retrieved on November 29, 2021.
The document is copyrighted. Any fair dealing for the purpose of private study or research cannot be reproduced without written permission. The content is not intended to be used for anything other than information purposes.
