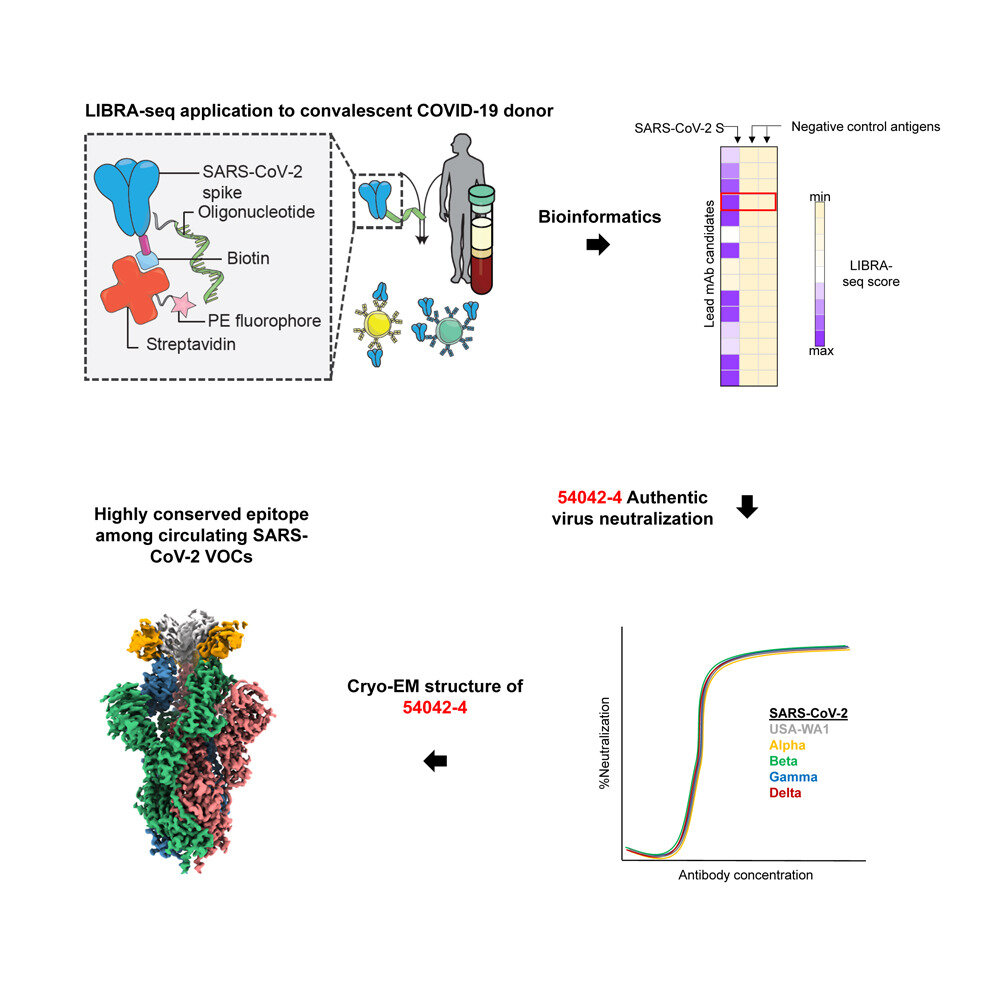
Graphical abstract. Credit: DOI: 10.1016/j.celrep.2021.109784
Vanderbilt University Medical Center developed a technology that allowed the discovery of an "ultrapotent" monoclonal antibody that can be used against multiple SARS-CoV-2 variants, including the Delta variant.
Researchers reported Sept. 15 that the antibody had rare properties, making it an important addition to the limited number of broadly reactive anti-body therapeutic candidates.
LIBRA-seq technology has made it possible to quickly identify antibodies that could neutralize SARS-CoV-2. Researchers can also screen for antibodies against viruses that may cause human disease, even though they have not yet been proven to be harmful.
"This is one of the best ways to prevent future outbreaks," said Ivelin Georgiev, Ph.D., director, Vanderbilt Program for Computational Microbiology and Immunology and associate Director of the Vanderbilt Institute for Infection, Immunology and Inflammation.
Georgiev, an associate professor of Microbiology, Pathology & Immunology and a member the Vanderbilt Vaccine Center, said, "The pathogens are constantly evolving, and it's basically playing catch up."
To prevent another COVID-19 outbreak, he suggested that a proactive approach is required to anticipate future outbreaks.
Georgiev and colleagues report on the isolation of a monoclonal anti-SARS-CoV-2 antibody from a patient with COVID-19. It is also effective against variants that slow down efforts to contain the pandemic.
This antibody is distinguished by its unique genetic and structural features, which distinguish it from monoclonal antibodies used to treat COVID-19. SARS-CoV-2 is expected to be less likely than other monoclonal antibodies to escape an antibody that it hasn’t seen before.
LIBRA-seq is for Linking B-cell receptor to antigen specificity via sequencing. It was created in 2019 by Ian Setliff (Ph.D.), a former student in Georgiev’s lab and now works in biotechnology. Andrea Shiakolas is a Vanderbilt graduate student.
Setliff wanted to know if it was possible to simultaneously map the genetic sequences and identities of specific viruses antigens. These are the proteins that antibodies recognize and attack. It was important to identify antibodies faster that can be targeted at a particular viral antigen.
Georgiev tested Setliff's idea with the support of VUMC's core genomes laboratory, Vanderbilt Technologies for Advanced Genomics, Vanderbilt Flow Cymetry Shared Resource and Vanderbilt University’s Advanced Computing Center for Research and Education. It worked.
Setliff's and Shiakolas' efforts culminated in a manuscript that described proof-of concept development of the LIBRASeq technology. It was published in Cell in 2019.
Georgiev stated that it would have been impossible to move at the pace we are now three or four years ago. "Much has happened in a short time, both in monoclonal antibody research and vaccine development.
There is no time to waste. He said that if the virus is given enough time, there will be many variants of the virus. One or more variants may even be worse than the delta variant.
Georgiev stated, "That's precisely why you should have as many options possible." "Basically, the antibody described in this article gives you another tool in your toolbox."
Continue exploring New tool could speed up antibody and vaccine research
More information: Kevin J. Kramer and colleagues, Potent neutralization SARS-CoV-2 variants by an antibody with an unusual genetic signature, structural mode of spike recognition, Cell Reports (2021). Information from Cell Reports, Cell Kevin J. Kramer, et al., Potent neutralization SARS-CoV-2 varieties of concern by an antibody that has an unusual genetic signature and a structural mode of spike recognition. (2021). DOI: 10.1016/j.celrep.2021.109784
