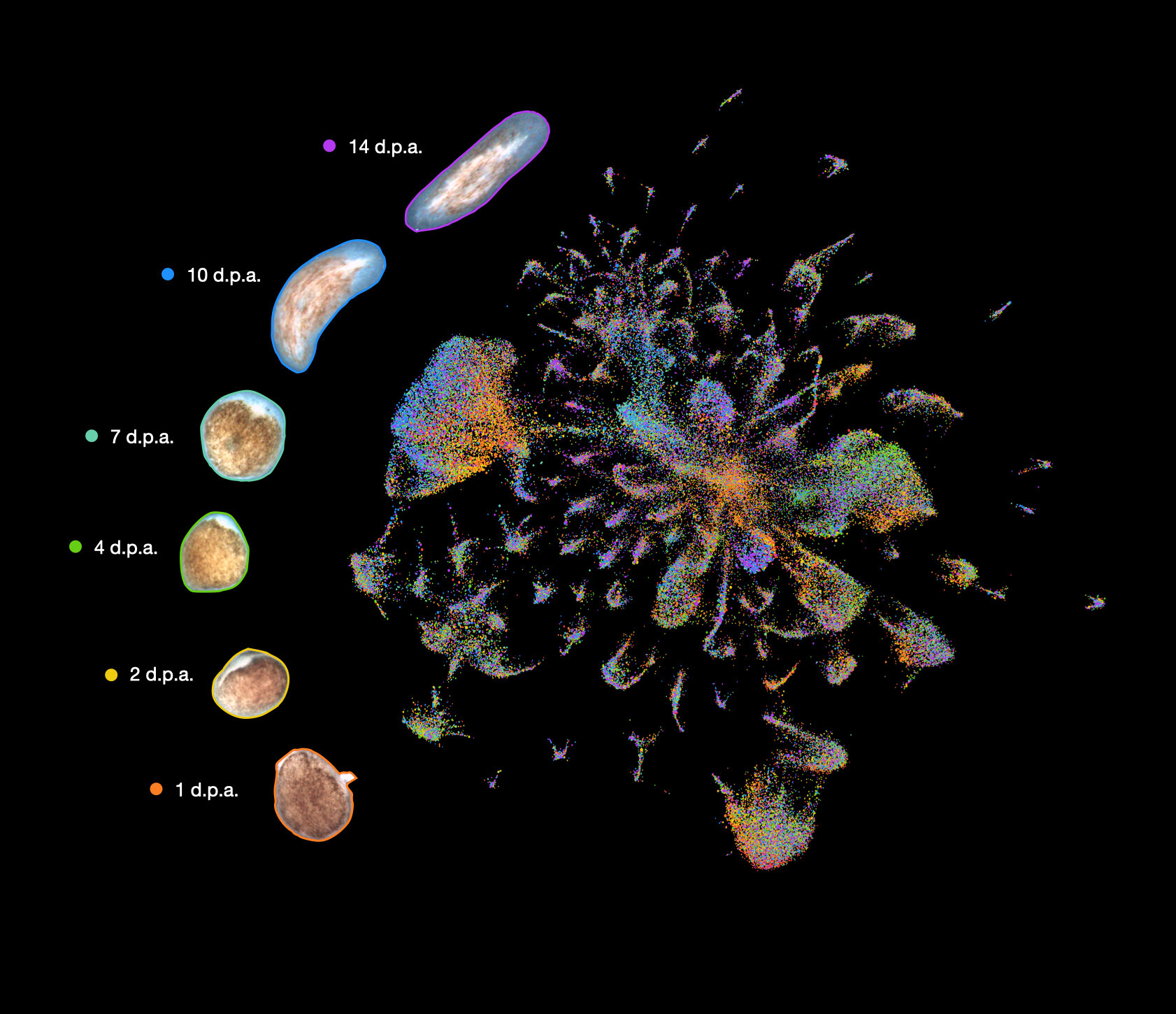
Flatworm regeneration is represented in an atlas. Each flatworm cell is represented as a dot, with colors representing collection times and distances representing similarities in gene expression profiles. Credit: Snchez Alvarado Lab
Fishing and using earthworms for bait is common. They can regenerate a head and tail if they are cut in half. It is impressive but not as easy to rehabilitate an earthworm than the planarian Schmidtea mediumea. This flatworm species can regenerate an entire animal using tiny tissue fragments that are as small as 1/279th.
This is how it happens. How do these cell types contribute to this amazing regenerative ability? What other types of cells are necessary for this amazing regenerative capacity, besides stem cells?
Recent research by the Snchez Alvarado Lab of the Stowers Institute for Medical Research has provided some answers to these complicated questions.
Blair Benham-Pyle (Lead Author), is a postdoctoral scientist at Stowers Institute and Chief Scientific Officer. He was also an investigator at Howard Hughes Medical Institute, Alejandro Snchez Alvarado.
Benham-Pyle says, "This is the first study to definitively find that all three germ layers of Schmidtea Mediterranea (muscle, epidermis and intestine), transcriptionally responds to amputation and that both tissues close to the wound site as well as far from it are contributing to the regenerative capacity."
"Regeneration was not a very well-known phenomenon before. We knew some important genes and could see how they were affected globally by amputation or during regeneration. But we didn't know how the behavior and function of individual cells across an animal. This experiment enabled us to identify that.
Benham-Pyle described it as "The dream experiment" and they were able to "characterize gene expression at the single-cell level across all of the cell types of a regenerating animal over time."
The researchers initially considered using large-scale single-cell sequencing to do the experiment. Droplet-based single-cell sequence, where every cell is enclosed in a droplet with a code and then lysed for labeling all mRNAs with the barcode, was not possible at the scale required for this experiment. Snchez Alvarado discovered a preprint posted to bioRxiv in 2017 that described a new single cell sequencing method called SplitSeq. After Benham-Pyle reviewed the preprint and had discussed the merits with Snchez, Alvarado, they decided to give it a try. After many tries, optimizations and troubleshooting with the molecular Biology and cytometry technology centers teams, Benham Pyle was able to bring a single-cell sequencing technology into the Stowers Institute.
Benham-Pyle, along with colleagues, captured nearly 300,000 single-cell transcriptomes from eight tissues and the stem cells compartment of animals that had lost their ability to regenerate. This was compared to animals that could regenerate.
Benham-Pyle explains, "This allowed us look at all the cell types across an animal to see which responded and what genes marked these cells as they changed or responded to regeneration."
Five cell types were identified and characterized by the researchers from each of the three germ layers. They had transiently altered transcription after an amputation. Benham-Pyle says that when genes from these cell types were removed, they found that each contributes to regeneration in different ways. They are activated at different times, in different parts, and at different times.
Some of their discoveries were more surprising than others. It was obvious, for example, that muscle is essential for patterning and that the epidermis plays a crucial role in early stem cell proliferation bursts during regenerative regeneration. The researchers were surprised, however, to discover rare cells, states induced during whole-body regeneration, called transient regeneration-activating cell states (TRACS), and to find that the intestine seems to be important for both stem cell maintenance and regulating tissue remodeling after amputation.
Benham-Pyle says, "I didn’t expect the intestinal system to change its output globally and remodel its function after an injury." It makes sense if you really think about it. Planarians usually grow their body plans based on the environment in which they live. The worm eats and this fuels stem cell proliferation and new biomass. The animal often becomes unable to eat if it is cut, particularly in severe injury. All the growth and remodeling must now be powered by the nutrients already present in the body. After an amputation, the function of the intestine is altered to remove material from the body and convert it into healthy cells in a regeneratedworm.
It took a team effort to acquire and make sense of the data.
Benham-Pyle explains that we had to complete all our manuscript revisions in the COVID-19 pandemic. This was when we were only at half of our research capacity. "Sean McKinney of the Microscopy Center discovered ways to automate imaging. We devised a system that allowed me to give them forty-eighty slides at a given time, with all kinds of samples and RNAi conditions. This was then imaged in overnight runs. The scanning confocal microscope was able to produce terabytes worth of imaging data, which gave us the boost we needed to accept the paper. They set the bar very high for microscopy facilities.
Carolyn E. Brewster is a bioinformatics specialist, who helped analyse the data and created the website. Aubrey M. Kent was also involved in the creation of the website. Frederick G. Mann, Ph.D. helped clone many genes Benham-Pyle screened for and characterized in this paper. Shiyuan Chen, Allison R. Scott, Andrew C. Box, Alejandro Snchez Alvarado and Alejandro Snchez Alvarado were other coauthors.
Benham-Pyle says that the paper takes a step back and "takes a global look" at what cells are needed to signal stem cells to stimulate them to create new tissue or replace lost tissue.
It turns out that many genes we have characterized, such as those in the intestine and the liver, are also implicated in immune evasion, which is a process that can lead to cancer or wound healing. Many of the same mechanisms stem cells use to bypass the immune system and fuel proliferation and growth during renewal may also be used by tumors. Understanding the non-stem cells and tissue types that help to create this signaling environment may lead to new targets to stimulate healthy wound healing in situations where regenerative capacity is limited or limit growth of things we don't want, such as tumors.
"Now that we have a map we can go and find out how cells talk to one another, what they are doing and how it's happening."
Continue reading Scientists develop the first map of human skeletal muscles development
More information: Identification of rare, transient post-mitotic cell states that are induced by injury and required for whole-body regeneration in Schmidtea mediterranea, www.nature.com/articles/s41556-021-00734-6 Journal information: Nature Cell Biology Identification of rare, transient post-mitotic cell states that are induced by injury and required for whole-body regeneration in Schmidtea mediterranea, DOI: 10.1038/s41556-021-00734-6
