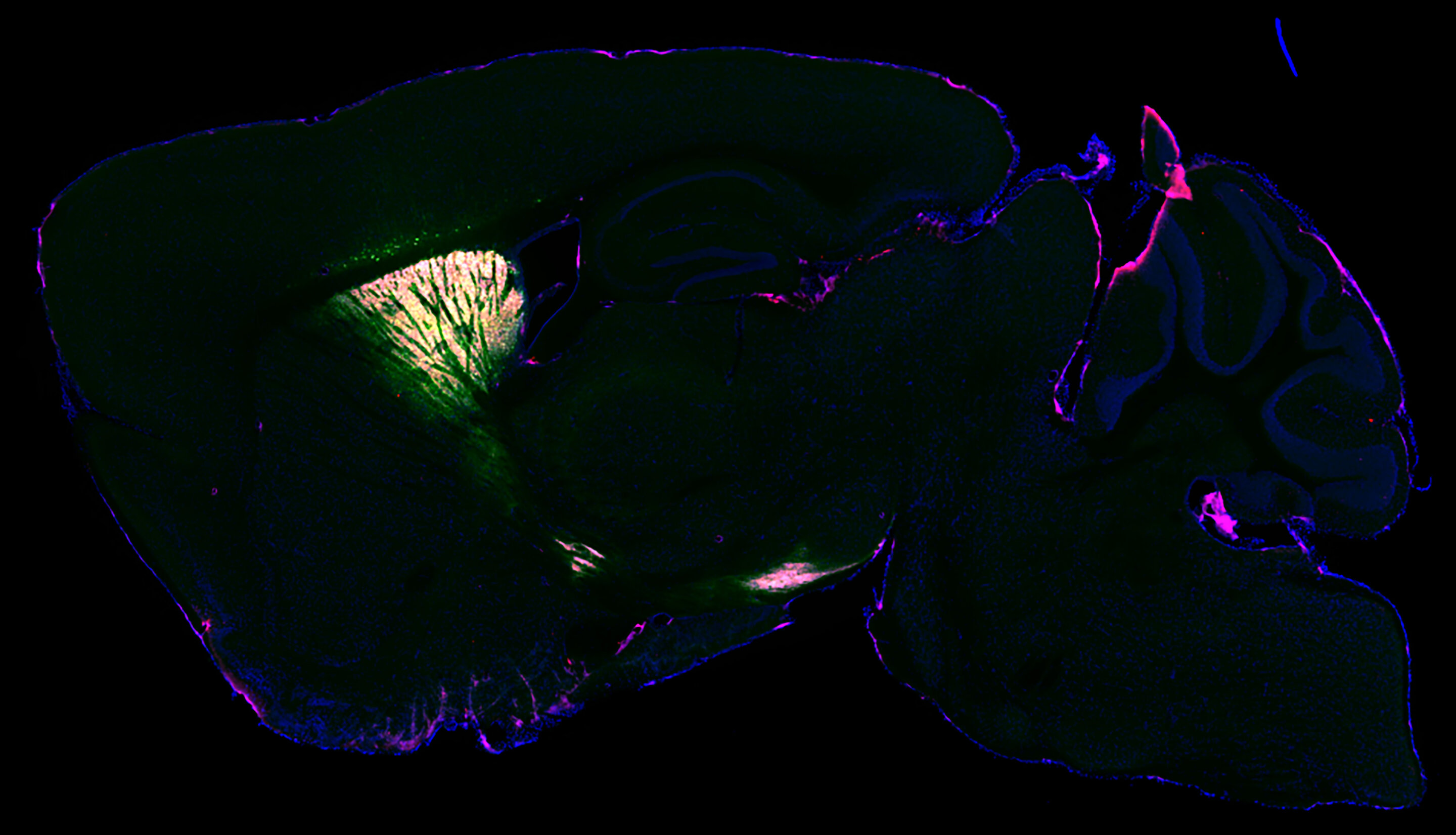
Vasin Dumrongprechachan, Ph.D. candidate, captures protein expression from the mouse brain to mass spectrometry analysis. This is visualized using fluorescence microscopy. Images show a cross section of the mouse's striatum. The outline of the brain is in blue. Magenta and green are the selectively tagged proteins that mass spectrometry analyses can identify. Credit: Northwestern UniversityResearchers have created a method that allows them to identify proteins in different types of neurons in the brains of living animals for the first time.The new study, led by Northwestern University and University of Pittsburgh is a huge step towards understanding the brain's many millions of proteins. Proteins are the building blocks of all cells, including neurons. They can help us understand complex brain diseases like Alzheimer's and Parkinson's. This could lead to new treatments.The study will appear in Nature Communications on August 11, 2012.Researchers created a virus that would send an enzyme to a specific location in the brains of living mice. The enzyme is derived from soybeans and genetically tags the proteins it binds to in a specific location. The researchers validated the technique by imaging the brain using electron microscopy and fluorescence. They found that their technique captured a snapshot of all the proteins (or proteomes) within living neurons. This can then be analysed postmortem using mass spectroscopy."Similar work was done in cell cultures before. However, cells in a dish don't work in the same way as they do in a human brain. Furthermore, they don't have identical proteins in the same places and do different things," Yevgenia Kozrovitskiy from Northwestern, who was the senior author of this study. It's much more difficult to do this work with complex tissue like a mouse brain. We can now take our proteomics prowess, and apply it to more realistic neural circuits that have excellent genetic traction.Researchers can now chemically tag proteins and their neighbours to see how they interact with each other in a proteome. Researchers used the virus that carried the soybean enzyme to create a separate green fluorescent protein.Kozorovitskiy stated that the virus acts as a message we send. "In this instance, the message contained a special soybean enzyme. We then sent a separate message with the green fluorescent protein. This would show us which neurons had been tagged. If the neurons turn green, we can tell that the soybean enzyme has been expressed in them.Kozorovitskiy, the Soretta and Henry Shapiro research professor of Molecular Biology, is an associate professor of neurobiology at Northwestern's Weinberg College of Arts and Sciences and a member of The Chemistry of Life Processes Institute. Matthew MacDonald was an assistant professor of psychotherapy at the University of Pittsburgh Medical Center. She led the project.Protein targeting plays catch-upGenetic targeting has revolutionized biology and neuroscience but protein targeting is still a far cry. Researchers can sequence and amplify genes to find their specific building blocks. However, proteins cannot be sequenced or amplified in the same way. Researchers must instead divide proteins into peptides, then put them back together. This is slow and inefficient.Kozorovitskiy stated that although we have gained a lot of momentum with genetic andRNA sequencing, proteins have been left out of the loop. But everyone understands the importance protein. Proteins are the ultimate effectsors of our cells. It is important to understand where proteins are located, how they function and how they relate to one another.Vasin Dumrongprechachan is a Ph.D. candidate at Kozorovitskiy’s laboratory and the paper’s first author. "Mass spectroscopy based proteomics [is] a powerful technique," he said. "Using our approach, it is possible to map the proteomes of different brain circuits with high precision. They can be quantified to determine how many proteins are found in each part of the brain and neurons.Next step: Better understanding brain diseasesThe new system is now validated and can be used by researchers to create mouse models of disease in order to better understand neurological disorders.Dumrongprechachan stated that they are looking to expand this approach in order to identify biochemical modifications to neuronal proteins during certain patterns of brain activity, or changes induced with neuroactive drugs to facilitate clinical advancements.Kozorovitskiy stated that "we look forward to applying this to models related brain diseases and connecting those studies to postmortem proteomics works in the human brain." It's ready for use in those models and we are eager to get started.Information: Nature Communications (2021) "Cell-type, subcellular compartment-specific APEX2 nearby labeling reveals activity dependent nuclear proteome dynamics within the striatum," Nature Communications (2021). Information from Nature Communications: "Cell-type proximity labeling reveals activity dependent nuclear proteome dynamics within the striatum" (2021). DOI: 10.1038/s41467-021-25144-y
