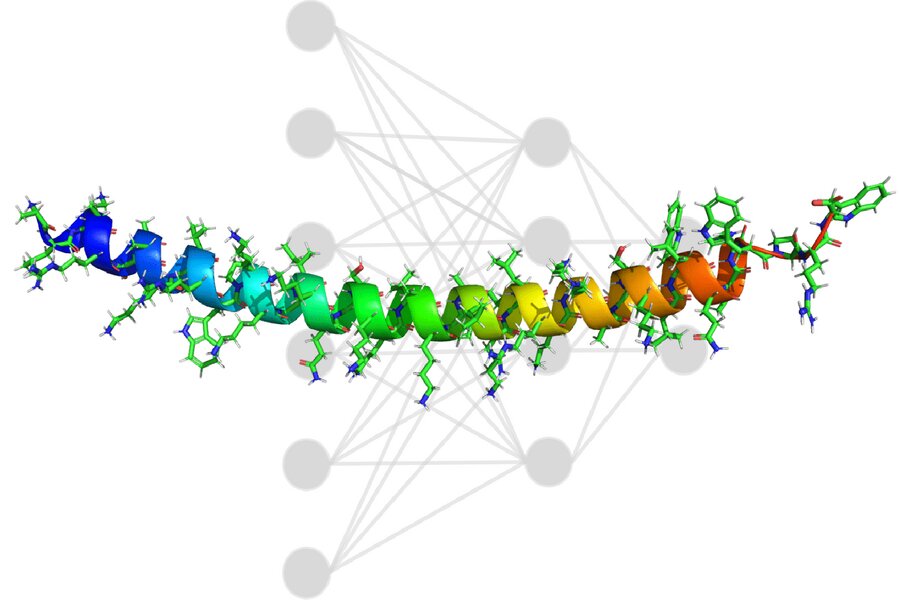
Researchers at MIT combined experimental chemistry and artificial intelligence to create non-toxic, highly active peptides that are compatible with phosphorodiamidate morpholino oligomers to facilitate drug delivery. Researchers hope to accelerate gene therapy development for Duchenne muscular disorder and other diseases by developing these new sequences. Credit: Massachusetts Institute of TechnologyDuchenne muscular disorder (DMD) is a rare genetic condition that affects young boys. It gradually weakens the muscles throughout the body, until the heart and lungs stop working. Symptoms usually appear by age 5. As the disease progresses patients begin to lose their ability to walk around the age of 12. The average DMD patient's life expectancy is now around 26.The announcement by Sarepta Therapeutics, a Cambridge, Massachusetts-based company, in 2019 of a drug that directly targets DMD's mutation was huge news. Antisense phosphorodiamidate-morpholino Oligomers (PMO) is the therapy. This large synthetic molecule permeates cells to modify the dystrophin genes, which allows for the production of a crucial protein that is missing in DMD patients. But PMO is not the only problem. It is not very good at entering cells," Carly Schissel (a Ph.D. student in MIT's Department of Chemistry) says.Researchers can attach cell-penetrating proteins (CPPs), to the drug to increase its delivery to the nucleus. This allows the drug to cross cells and reach the target. It is still a mystery which peptide sequence is most effective for this job.MIT researchers now have a system for solving this problem. They combine experimental chemistry and artificial intelligence to find nontoxic, highly active peptides that are easily attached to PMO to facilitate delivery. They hope to accelerate gene therapy development for DMD and other conditions by developing these new sequences.The results of the study were published in Nature Chemistry. They are led by Schissel and Somesh Mhapatra, a Ph.D. student at the MIT Department of Materials Science and Engineering. Bradley Pentelute (professor of chemistry) and Rafael GomezBombarelli (assistant professor of materials science, engineering), are the senior authors of this paper. Justin Wolfe and Colin Fadzen are also authors."Proposing new propeptides using a computer is easy. Gomez-Bombarelli says that the hardest part is judging if they are good or not. "The key innovation is using machine learning to connect the sequence of a peptide, particularly a peptide that includes non-natural amino acids, to experimentally-measured biological activity."Data from the futureCPPs are short chains that contain between five to 20 amino acids. Although one CPP may have a positive effect on drug delivery, many CPPs can be linked together to have a synergistic effect when carrying drugs to the end. These shorter chains, which contain 30 to 80 amino acid, are known as miniproteins.Researchers on the experimental side need to build a solid dataset before a model can make any useful predictions. Schissel and her collaborators were able to create a library of 600 miniproteins each, each with a PMO. The team was able, using an assay to determine how each miniprotein could move their cargo around the cell.It was crucial to determine the activity of each sequence with PMO attached. It is likely that any drug will alter the activity of a CPP-sequence. Therefore, it is difficult for existing data to be repurposed. Data generated in one lab on the same machines by the same people meet a gold standard in consistency in machine learning datasets.The project had one goal: to develop a model that can be used with any amino acid. There are only 20 amino acids that naturally occur in the body. However, there are hundreds more available elsewhere. Researchers use one-hot encoders to represent them in machine-learning models. This is a method that assigns each component of the model to a set of binary variables. For example, three amino acids would be represented by 100, 010 and 001. Researchers would have to add more variables to be able to add new amino acids.Instead, the team decided to represent amino acids using topological fingerprinting. This is basically creating a unique barcode that represents each sequence. Each line of the barcode indicates whether a specific molecular substructure is present or absent. Mohapatra, who was responsible for the project's development, said, "Even though the model has never seen [a sequence], we can represent it in a barcode which is consistent with what the model has seen." Researchers were able expand their repertoire of sequences by using this representation system.The convolutional neural network was trained on the miniprotein library by the team. Each of the 600 miniproteins was labeled with the activity to indicate its ability to penetrate the cell. The model suggested miniproteins laden in arginine. This amino acid can cause a hole to the cell membrane and is not good for keeping cells alive. Researchers used an optimizer in order to decentivize the arginine. This solved the problem and prevented the model from cheating.The model's ability to interpret the predictions made by it was crucial. Gomez-Bombarelli states that a black box is often not sufficient because models might be focusing on an incorrect phenomenon or could not exploit it correctly.Researchers could use this method to overlay the predictions of the model with sequence structure barcodes. Schissel says that this highlights the regions where the model believes play the most important role in high activity. It's not perfect but it provides you with specific regions to work with. This information will be very useful in the future when we try to create new sequences empirically.Delivery boostThe machine-learning model suggested sequences that were better than any other known. One sequence in particular can increase PMO delivery 50-fold. The researchers confirmed their predictions by injecting mice with the sequences suggested by computers. They also proved that the miniproteins were safe for humans.Although it is still too early to predict how this will impact patients, better PMO delivery will make a difference in many ways. Patients may have fewer side effects or need to take fewer doses of PMO (PMO is intravenously administered, usually on a weekly basis). It may also be less expensive. Recent clinical trials have shown that Sarepta Therapeutics' proprietary CPP could reduce PMO exposure by 10 times. Miniproteins can also be used to improve PMO. Additional experiments revealed that the miniproteins generated by the model carried functional proteins into cells.Mohapatra noticed a gap between machine-learning researchers' work and that of experimental chemists. He posted the model to GitHub along with a tutorial, which Mohapatra shared with experimentalists who own their own sequences and activities. Mohapatra notes that the model has been adopted by more than a dozen people around the globe, who have repurposed it to create their own powerful predictions for a variety of drugs.Learn more about personalizing cancer treatment using machine learningFurther information: Carly K. Schissel and colleagues, Deep learning to create nuclear-targeting miniproteins for abiotics, Nature Chemistry (2021). Information from Nature Chemistry Carly K. Schissel and colleagues, Deep learning to create nuclear-targeting miniproteins for abiotics, (2021). DOI: 10.1038/s41557-021-00766-3This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.
