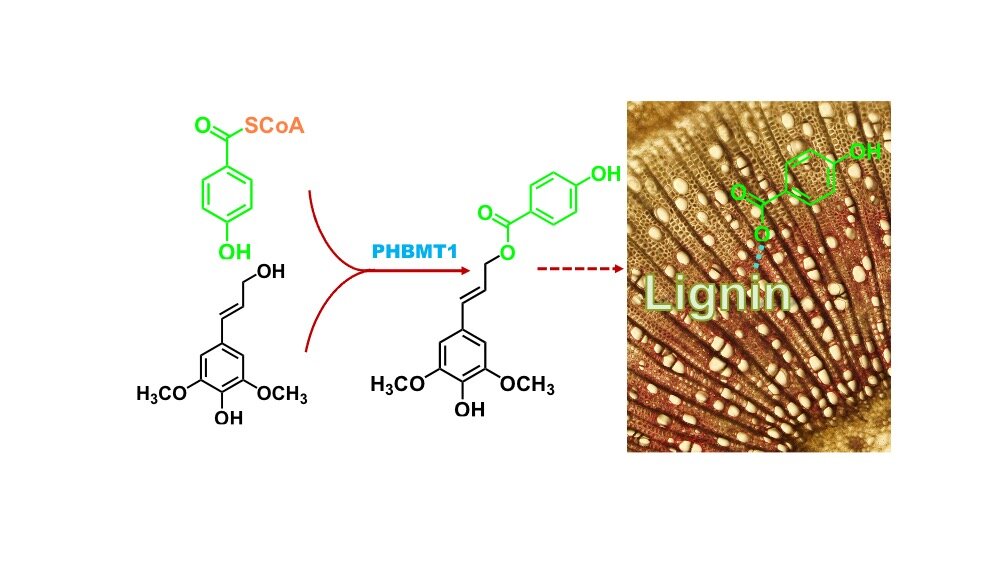
Brookhaven Lab scientists discovered an enzyme (PHBMT1) which transfers p-hydoxybenzoate, (green), to poplar lignin building blocks. The resulting conjugate is then incorporated into the nascent lignin polymer, leading to p-hydoxybenzoate-decorated lignin. Scientists may be able to engineer plants to produce more of this critical industrial chemical building block. Credit: Brookhaven National LaboratoryScientists have discovered an enzyme that can be used to make a major industrial chemical from woody poplar trees by studying the biochemistry and cell walls of plants. Nature Plants published the research. It could open up a sustainable route for "p-hydroxybenzoic Acid," a chemical building block that is currently made from fossil fuels in plant biomass.P-hydroxybenzoic Acid is a versatile chemical feedstock. It can be used as a building block to make liquid crystals, a plasticizer of nylon, a sensitizer of thermal paper, and a raw materials for making parabens, dyes and pigments," Chang-Jun Liu (a U.S. Department of Energy plant biochemist and the lead author of the paper) said.Global market value for p-hydroxybenzoic acids was $59 million in 2020. It is expected to rise to $80 million by 2026. The current process of making this crucial chemical is dependent on petrochemicals. The harsh reactions required for its synthesis (high pressure and high temperature) have negative environmental consequences. It is possible to create p-hydroxybenzoic acids in plants economically and sustainably. This could reduce environmental impacts and help to build a bioeconomy.Liu said that he had identified a key enzyme that is responsible for the synthesis of and the accumulation of p–hydroxybenzoate, (pBA), the conjugate base of the p-hydroxybenzoic acidsin lignin. This polymer is one of the three major ones that makes up the structural support surrounding plant cells. This discovery could allow us to engineer plants to increase the amount of this chemical building block within their cell walls and potentially add value to the biomass.Biofuels, bioproductsCell walls are composed of a mixture of chainlike polymerscellulose and hemicellulose. These are the main sources of plant biomass. Liu and other scientists are studying the biochemical pathways used to make these plant polymers. One of the goals was to discover how changing the polymers can make it more economical and easier to convert biomass into biofuels.Lignin is a crucial component of plants' structural integrity, mechanical strength and waterproofing. It can be difficult to remove. Recent research has led to technological advances that have opened up new opportunities for increasing the use and value of lignin.Scientists know that sidechains are attached to lignin building blocks. These side groups have a function that is not clear. Liu's group was keen to explore their impact on lignin structure, properties and behavior. They set out to find the enzyme that attaches pBA to lightenin.Liu stated that if we could identify the enzyme and control its expression, we could control the amount of pBA found in bioenergy plants' biomass.Looking for the geneTheir study was conducted on poplar. The poplar tree is a fast-growing species with rich woody biomass. It is a promising renewable source of feedstock for biofuel production and bio-based chemicals. It also uses pBA for its main sidechain "decoration".Liu's group screened a number of candidate genes from poplar to identify the enzymes involved in attaching pBA and other chemical groups lignin."We cloned twenty candidate genes that are primarily expressed within woody tissues. They encode enzymes called Acyltransferases. These enzymes are most likely to transfer chemical groups to specific accepter molecules," Liu stated.Scientists mutated the genes to create enzymes and then mixed them with different building blocks, including one carbon compound that was isotope-labeled. Scientists were able to trace the isotope label as well as other biomolecular techniques using test tubes, which allowed them to determine whether any candidate enzyme was involved with attaching sidechains like pBA (or other chemical groups). They were able narrow down the most likely candidate to cause the reaction of interest.It was difficult to prove the enzyme's functionality in plants. It took many years for scientists to accomplish this task, and new developments in molecular biology were needed.CRISPR/Cas9 is a new "genetic scissor", which allows precise editing of the genes of target organisms. CRISPR/Cas9 was used by the team to create a poplar variety in which the candidate enzyme-encoding genes had been deleted. The subsequent analysis revealed that these plants had almost no pBA in their stems.Another genetic test was also performed by the overexpression of the gene that makes the candidate enzyme. The plants with higher levels of pBA exhibited increased growth.Liu stated that these data are conclusive evidence that the gene/enzyme identified can attach the lignin building block pBA together.One way to produce p-hydroxybenzoic acids sustainably is to increase the pBA content of plants through genetic manipulation.Researchers also discovered that lignin from plants with lower pBA was easier to dissolve in solvents. This suggests that pBA in nature helps to strengthen lignin.Another outcome of the identification of the enzyme to add pBA lignin to could be genetic strategies to tailor the chemical properties of the lignin.A lower pBA could improve the "delignification” of woody biomass in processes like pulping, papermaking, and biofuel production.However, increasing the pBA level of lignin could increase timber durability and provide a pathway to long-term carbon sequestration through locking up more carbon in plant biomass. This is another key DOE goal.John Shanklin (Chair of Brookhaven Lab Biology Department) said, "This work is an excellent example of basic scientific research leading into potentially valuable downstream applications."Continue reading: A new enzyme can break down waste to make biofuels and bioproducts that are less expensiveMore information: Monolignol acyltransferase for lignin p-hydroxybenzoylation in Populus, Nature Plants (2021). www.nature.com/articles/s41477-021-00975-1 Journal information: Nature Plants Monolignol acyltransferase for lignin p-hydroxybenzoylation in Populus,(2021). DOI: 10.1038/s41477-021-00975-1
