Cells are smaller than a grain of salt and make up all life. Their structures look simple, but they are complex and allow them to carry out the functions that sustain life.
Researchers are starting to be able to see this activity in a more detailed way.
If you start at the level of the whole organisms and work down, or if you start at the level of single atoms and work up, you can see a biological structure.
There is a resolution gap between the smallest and largest structures of a cell.
Scientists have been able to see entire cities and individual houses, but they didn't have the tools to see how the houses came together to make up neighborhoods.
Understanding how individual components work together in a cell is dependent on seeing the neighborhood-level details.
The gap is being bridged by new tools. The development of one particular technique has the potential to deepen how researchers study and understand how cells function.
As the former editor-in-chief of Science magazine and as a researcher, I have seen amazing progress in the development of tools that can determine biological structures in detail.
Understanding how biological structures fit together in a cell is important to understanding how organisms function.

Light microscopy showed the existence of cells. In the 20th century, electron microscopy offered even greater detail, revealing the elaborate structures within cells, including the endoplasmic reticulum.
In the 1940s to 1960s, biochemists separated cells into their components and learned how to determine the 3D structures of the macromolecules. X-ray crystallography was used to see the structure of myoglobin.
Nuclear magnetic resonance, which produces images based on how atoms interact in a magnetic field, has increased the number and complexity of structures scientists can see.
A camera can be used to detect how a beam of electrons is traveling through a sample.
The samples are quickly frozen to prevent them from being damaged by radiation. A detailed model of the structure of interest can be created by taking multiple images of individual molecule and averaging them into a 3D structure.
Similar parts are shared with cryo-EM, but different methods are used. A region of interest in a cell can be thinned by using an ion beam.
The sample is tilted to take multiple pictures of it at different angles, similar to aCT scans of a body part. 3D images of a portion of the cell are produced by combining the images.
Researchers can identify the components of a cell with the high resolution of this image. This approach has been used by researchers to show how a cell degrades its own genes.
Many of the steps researchers once had to do manually to determine the structures of cells are becoming automated.
It is possible to combine artificial intelligence programs like AlphaFold with cryo-EM to help interpret images.
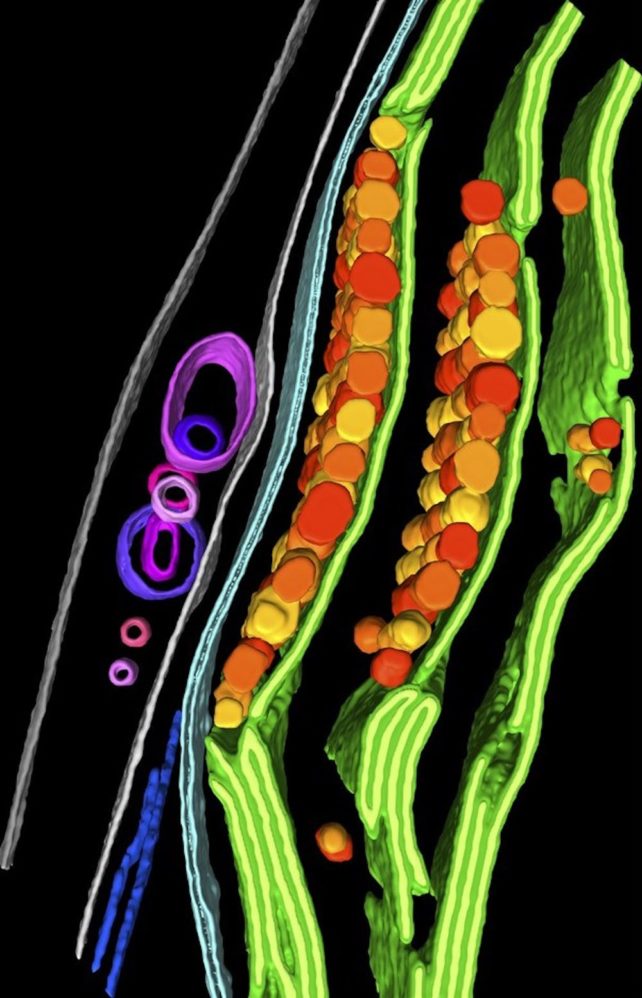
Researchers will be able to tackle some key questions in cell biology with different strategies because of the improved techniques.
The first thing to do is to figure out which parts of the cells to look at. A visualization technique called correlated light and electron microscopy uses fluorescent tags to help locate regions where interesting processes are taking place.
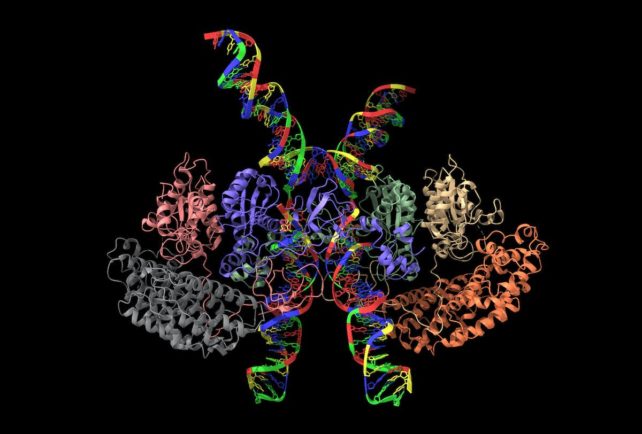
Additional insight can be provided by comparing the genetics of cells. Scientists can look at cells that can't do certain things and see how their structure reflects this. Researchers can use this approach to study how cells communicate.
The tool is likely to stay a specialized one. Increased accessibility and technological developments will allow the scientific community to look at the link between cellular structure and function at previously unavailable levels.
New theories on how we understand cells will move from disorganized bags of molecule to intricately organized and dynamic systems.
Jeremy Berg is an associate senior vice chancellor for science strategy and planning at the University of Pittsburgh.
Under a Creative Commons license, this article is re-posted. The original article is worth a read.