Simulations can take weeks or months to run. There is a new approach to epidemic modeling in a recent study.
The study uses sparsification, a method from graph theory and computer science, to find the most important links in a network.
By focusing on critical links, the authors were able to reduce the computation time for simulation of the spread of diseases through highly complex social networks.
"Epidemic simulations require substantial computational resources and time to run, which means your results might be outdated by the time you are ready to publish," says Alexander Mercier, a graduate student at the Harvard T.H. "Our research could eventually enable us to use more complex models and larger data sets while still acting on a reasonable timescale when simulating the spread of Pandemics."
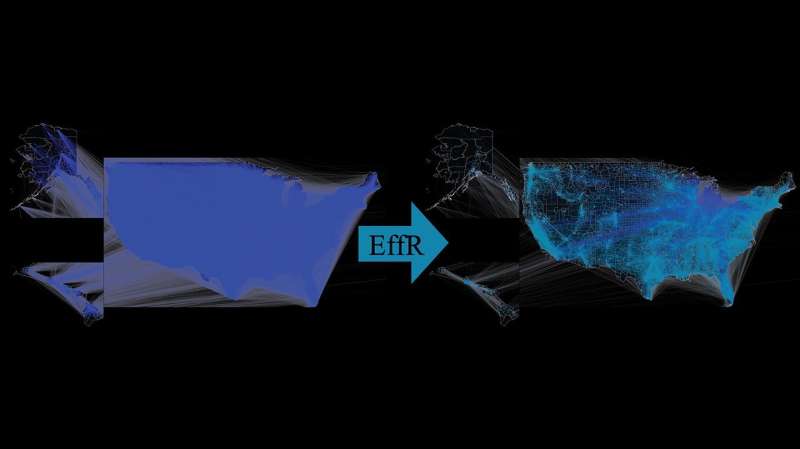
The data from the U.S. Census Bureau was used by the team to create a mobility network.
They applied several different sparsification methods to see if they could reduce the network's density while retaining its dynamics.
Effective resistance was the most successful sparsification technique. The total resistance between two endpoints in an electrical circuit is the basis of this technique. In the new study, effective resistance works by ignoring links that can be easily bypassed in favor of links that are the most likely avenues of disease transmission.
In the life sciences, low-weight links in a network are assumed to have a small chance of spreading a disease. Even a low-weight link can be important in an epidemic if it connects two distant regions.
The researchers created a network with 25 million fewer edges than the original U.S. commute network.
Moore says that the discovery that sparsification works for both linear and non- linear problems was a surprise.
While still in an early stage of development, the research not only helps reduce the computational cost of simulations of large-scale pandemics but also preserves important details about disease spread, such as the probability of a specific census tract getting infections and when the epidemic is likely to arrive there
Alexander Mercier and his co-authors wrote Effective resistance against pandemics: Mobility network sparsification for high fidelity epidemic simulations. There is a journal called the Journal of the PCbi.
Journal information: PLoS Computational Biology