Thanks to innovations in expansion microscopy, we can now see the inside of cells. Future insight into neuroscience, pathology, and many other fields could be provided by the advancement.
The paper "Magnify is a universalmolecular anchoring strategy for expansion microscopy" was published in the journal Nature Biotechnology.
The Associate Professor of Biological Sciences said that magnification can be a potent and accessible tool.
The Biophotonics Lab is a leader in the field of enabling super-resolution images of biological samples. Through the process, samples are embedded in a swellable hydrogel that expands to allow them to be seen. It is now possible to see the structures using standard microscopes, instead of using expensive high-resolution images.
Magnify is a variant of expansion microscopy that allows researchers to use a new formula that retains a spectrum of biomolecules, offers a broader application, and increases the expansion rate up to 11 times linearly or 1.
Some of the challenges of expansion microscopy were overcome by us. The universal strategy to keep the tissue's biomolecules in the expanded sample is one of the main selling points.
Many biomolecules that held tissues together were eliminated in previous protocols. Valuable information could be contained in these molecule.
He said that in the past, to make cells really expand, you needed to use enzymes to digest proteins, so in the end, you had an empty gel with labels that indicated the location of theProtein of interest Multiple biomolecules can be labeled in a single sample with the new method.
It was similar to having single choice questions. The version one protocol is the one you want to use. It would be a different version if you wanted to label the nucleus. It wasn't easy if you wanted to do simultaneous scans. You can label multiple items with the help of Magnify, such asProteins, Cholesterol and Carbohydrates.
The first co-authors of the paper were the lab researchers.
It is possible to image specimen in high resolution with this method. You need a lot of equipment and training. This method is applicable to many types of sample preparations and can be seen with standard microscopes in a biology lab.
The goal was to make the protocols compatible for researchers who would benefit from using the Magnify as part of their tool kits.
One of the ideas that we tried to keep in mind was to meet researchers where they are and have them change as few things as possible. It works with tissues that have been preserved and stored. It is very flexible in that you don't necessarily need to redesign to work with what you already have.
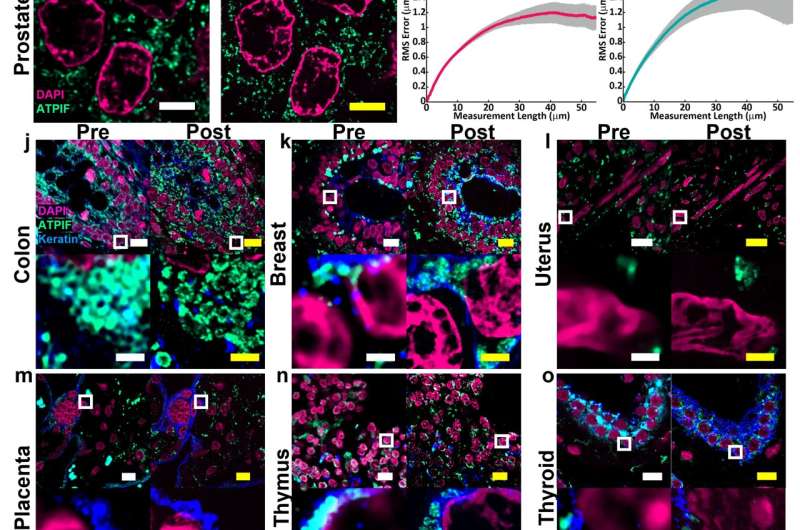
The fact that the new protocol is compatible with a wide range of tissue types is important for researchers such as Simon Watkins. Most expansion microscopy methods are designed to work in the brain. It was tested on samples from various human organs and tumors.
Watkins said that if you have a tissue with dense and non-dense components, this will get around tissues that previously wouldn't expand. Leon has been working on making this protocol work with tissues that have been preserved.
The lung tissue is studied by an assistant professor of biomedical engineering. He's researching the motile cilia that help clear mucus in the airway. The structures are too small to be seen without the use of electron microscopes. Ren's team collaborated with Zhao's lab to develop lung organoid models with specific defects in cilia that could be used to show clinically relevant cilia pathology.
"With the latest Magnify techniques, we can expand those lung tissues and start to see some ultrastructure of the motile cilia even with a regular microscope, and this will expedite both basic and clinical investigations" he stated.
The researchers were able to see the defects in the lung cells.
The lung tissue engineering community needs a better way to describe the tissue system that they work with. He hopes the work will be refined and applied to pathology samples found in tissue banks.
The hydrogel developed in the Zhao lab is more robust than its predecessors, which caused breaks during the process.
He said that they are hoping to make it more accessible to the community. There are many different directions this can take. A lot of people are interested in using this kind of technology for basic science.
Alison Barth is a professor of life sciences at Carnegie Mellon. She said that the new methods will benefit researchers.
Barth said that the brain is a great place to use these techniques. It will be beneficial to use microscope methods for synaptic phenotyping and analysis.
The method is able to work on many different types of tissue.
Other study authors include Piyumi Wijesekara and Emma F. DiBernardo.
There is a universal strategy for expansion microscopy. www.nature.com/articles/s4158
Journal information: Nature Biotechnology