A new method can illuminate the identities and activities of cells throughout an organ or a tumor at unprecedented resolution according to a study.
The method, described in a paper in Nature Biotechnology, records gene activity patterns and the presence of key proteins in cells while retaining information about the cells' precise locations. The creation of complex, data-rich "maps" of organs could be useful in basic and clinical research.
The technology allows us to map the spatial organization of tissues, including cell types, cell activities and cell-to-cell interactions, as never before.
10x Genomics is a California-based company that makes laboratory equipment for the profiling of cells in tissue samples. The three co-first authors were a researcher, a student, and a technician in the lab.
Scientists and engineers are trying to develop better ways to see how organs and tissues work. Recent years have seen the advancement of techniques for profiling genes and other information in individual cells. Information about profiled cells' original locations within the tissues is lost when the tissues are dissolved and the cells are separated. The new method captures that spatial information as well.
TheSPOTS method is based on 10x Genomics technology. It uses glass slides that can be used with normal microscope-based pathology methods, but are also coated with thousands of special probe molecule. Each probe molecule has a bar code on it's slide. When a thinly sliced tissue sample is placed on the slide and its cells are made permeable, the probe molecule on the slide grab adjacent cells' messengerRNAs, which are essentially the transcripts of active genes. The method includes the use of designer antibodies that bind to the proteins of interest in the tissue, as well as the special probe molecule. With swift, automated techniques, researchers can identify the captured mRNAs, and map them to their original locations. The maps can be compared to standard pathology images.
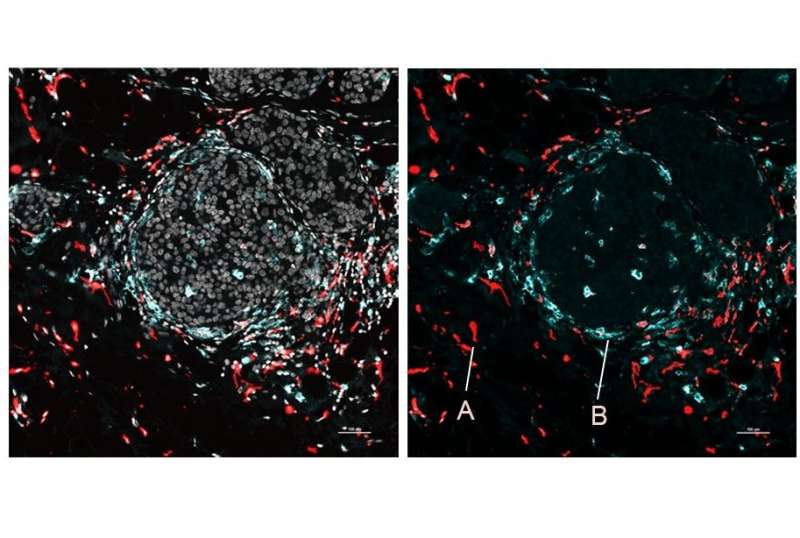
The team demonstrated SPOTS on tissue from a normal mouse spleen, showing the complex functional architecture of the organ, including clusters of different cell types, their functional states, and how those states differed with the cells' locations.
The investigators used SPOTS to map the organization of a mouse breast tumors. The map depicts two different states of the immune cells, one active and the other suppressing the immune system and forming a barrier to protect the tumors.
The difference in the microenvironment is likely driving the activity states of the two macrophage subsets.
Details of the tumor immune environment, which can't be resolved due to immune cells' sparseness within tumors, might help explain why some patients respond to immune- boosting therapy and some don't.
The initial version of SPOTS has a spatial resolution that sums data for at least several cells. The researchers hope to narrow this resolution to single cells and add other layers of cellular information.
There is more information about the integration of whole transcriptome spatial profiling. www.nature.com/articles/s4158
Journal information: Nature Biotechnology