The better we understand cellular processes, the better we can develop drugs. It has been difficult to track the regulation of non-codingRNA, which is not further converted into a transcript.
A research team from the Technical University of Munich has developed a reporter system that can be used to monitor the production of both coding and non- codingRNA.
For cellular processes, our genetic DNA information is transcribed intoRNA, which then undergoes further processing before it either serves as a blueprints for proteins or performs a cellular function itself A lot of the condition of our cells is revealed by the quantities of the different types ofRNA produced. In case of an infectious disease, cells make more of the molecule that codes for the immune response.
Existing reporter systems can be used to track the process of translation. Not all genes have the same function. Human genes are non-coding and include genes for long non-codingRNAs. These areRNAs with more than 200 building blocks that don't work as blueprints. Important processes are controlled by them. According to initial research, lncRNA is involved in a number of processes, such as the organization of structures in the cell nucleus or the switch on and off of certain genes.
It has been hard to investigate lncRNAs with existing methods. Classical reporter systems can't be used in fixed cells because they can't translate.
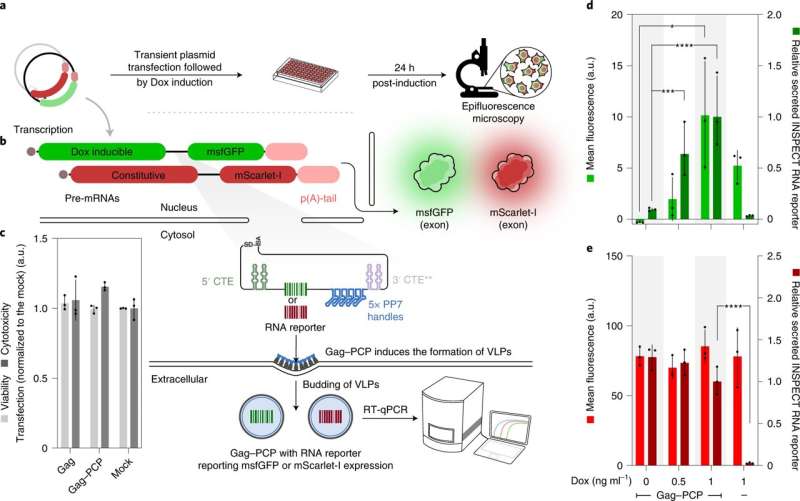
Non-codingRNA can be monitored by InSPECT.
InSPECT is a new reporter system. The newly developed reporter system was published in the journal Nature Cell Biology.
InSPECT uses modified introns to sequence reporter genes. The pre-matureRNA molecule is naturally removed by the cell during processing. The introns are transported to the cell's cytoplasm, where they are translated into reporter proteins, instead of being degraded after removal. Conventional methods can be used by the researchers.
InSPECT doesn't modify the completedRNA or the proteins.
The problem of tracking the generation of non-codingRNA is solved by the new tool. The reporter systems have to be fused directly to the RNA being studied in order to be co-translated. INSPECT modifies the introns.
The team has demonstrated the function of INSPECT with various examples. Interleukin 2 is produced in larger quantities in response to infections and was tracked by them. Highly sensitive monitoring of the production of two lncRNAs has been achieved by them.
InSPECT adds an important tool to the toolkit. Prof. Westmeyer says it makes it easier to study the role of non-codingRNAs in cell development and how they can be altered to prevent them from turning into cancer cells.
It may be possible in the future to study an entire genetic regulation process fromRNA processing to the production of specificprotein variant in living cells, if we combine the minimally-invasive reporter system EXSISERS.
There is more information about the Intron-encoded cistronic transcripts for minimally invasive monitoring of coding and non-codingRNAs by the authors.
Nature Cell Biology is a book written by Jeffery Truong and his co-authors. The DOI is 10.1038/s41556-021-0
Journal information: Nature Cell Biology