
The pearls that are sought after for necklaces, earrings, and rings are produced by pearl oysters. The industry brought in around 88 billion dollars in the early 1990s. The production of Japan's pearls has plummeted in the last 20 years due to diseases and red tides.
Researchers from the Okinawa Institute of Science and Technology collaborated with other research institutions to create a high-quality genome. The research was published in a scientific journal.
One of the authors said it was important to establish the genome. Many of the genes that are essential for survival are in the genes. We can do a lot of experiments with the complete gene sequence.
One of the first genomes assembled of a mollusk was published in 2012 by Dr. Takeuchi and his team. They continued genome-sequencing in order to establish a higher quality.
The oyster's genome is made up of 14 pairs of chromosomes and one set from each parent. There are almost identical genes on the two chromosomes of each pair.
Researchers usually combine the pair of chromosomes when they sequence a genomes. It works well for laboratory animals, which usually have the same genetic information. This method leads to a loss of information when it comes to wild animals.
The researchers decided against merging the chromosomes. The method that they used to sequence both sets of chromosomes is very rare. It's probably the first research that uses this method to focus on marine animals.
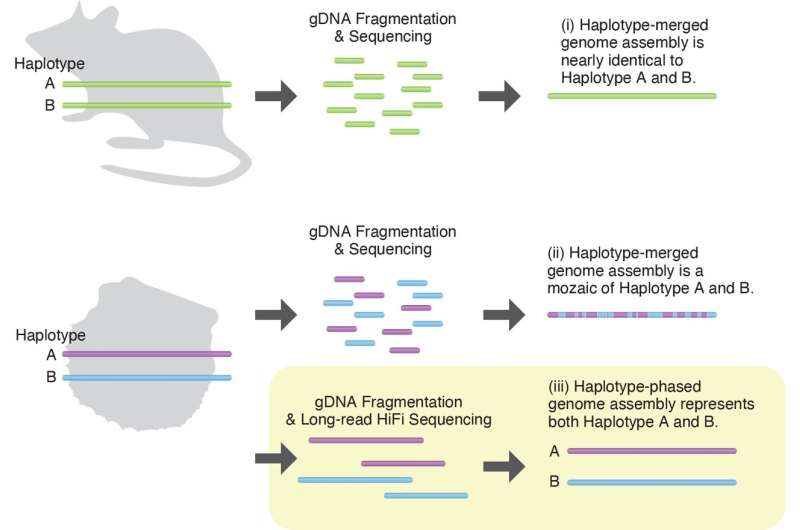
Pearls have 28 pairs of chromosomes. State-of-the-art technology was used to sequence the genome. One of the first authors was Dr. Takeuchi, who is now at the University of Tokyo. Many of the genes were linked to immunity.
There are different genes on a pair of chromosomes that can be used to identify infectious diseases.
He pointed out that when an animal is cultured, it can produce more beautiful pearls. The farmers breed two animals with this strain and that leads to inbreeding. After three inbreeding cycles, the genetic diversity was reduced. The immunity of the animal could be affected by reduced diversity in the chromosomes.
It is important to keep the genome diversity in the population.
A high quality haplotype-phased genome reconstruction reveals unexpected haplotype diversity in a pearl oyster. There is a book titled "10093/dnares/dsac035."