The startup behind Stable Diffusion is funding an effort to apply artificial intelligence to the frontiers of pharma. The first projects of the endeavor will be based on machine learning and genetics.
The company aims to explore the intersection of artificial intelligence and biology in a setting where students, professionals and researchers can work together.
According to Mostaque, Stability supports one of the independent research communities that is called Open BioML. We see an opportunity to advance the state of the art in sciences, health and medicine through OpenbioML.
One might be wary of Stability AI's first venture into healthcare given the controversy surrounding Stable Diffusion. The startup has taken a laissez-faire approach to governance, allowing developers to use the system as they please.
Machine learning in medicine is a difficult field to navigate. The tech has been used to diagnose conditions like skin and eye diseases, but research shows that it can lead to worse care for some patients. Statistical models used to predict suicide risk in mental health patients performed better for white and Asian patients than they did for black patients.
It was wise to start with safer territory for OpenbioML. First projects are:
Each project is led by independent researchers, but Stability AI is providing support in the form of access to itsAWS-hosted cluster of over 5000 Nvidia A 100 graphics cards to train the artificial intelligence systems. This will be enough processing power and storage to train up to 10 different AlphaFold 2-like systems in parallel according to Niccol.
Computational biology research leads to open- source releases. A lot of it happens at the level of a single lab and is constrained by insufficient computational resources. In order to change this, we want to encourage large-scale collaborations and back those collaborations with resources that only the largest industrial laboratories have access to.
Luca Pinello, a pathology professor at the Massachusetts General Hospital and Harvard Medical School, is the leader of the most ambitious project of OpenbioML. The goal is to use generative artificial intelligence systems to learn and apply the rules of the genes that are regulated. Misregulated genes are the cause of many diseases and disorders, but science has yet to discover a reliable process for identifying these regulatory sequences.
A type of artificial intelligence called a diffusion model can be used to generate cell-type specific regulatory DNA sequence. New data can be created by learning how to destroy and recover old data. The models get better at recovering data when they are fed samples.

The image is from OpenbioML.
Computational biology is one of the areas where fusion is starting to be applied. The application of DNA-Diffusion is currently being explored.
The DNA-Diffusion project will produce a model that can be used to generate regulatory DNA from text instructions. Improving the scientific community's understanding of the role of regulatory sequence in different diseases is one of the benefits of a model like this.
This is mostly theoretical. It's very early days, but the push to involve the widerai community is due to the promising preliminary research.
It is more likely to bear fruit when it is smaller. The goal of the project is to arrive at a better understanding of machine learning systems that predict the structure of the human body.
In his piece about DeepMind's work on AlphaFold 2, my colleague Devin Coldewey mentioned that the systems that accurately predict the shape of the human body are new on the scene. Different tasks are accomplished within living organisms with the help of different types of proteins. The process of determining the shape of an acids sequence used to be difficult and error prone. Thanks to AlphaFold 2 and other artificial intelligence systems, 98% of the human body's structure is known to science, as well as hundreds of thousands of other structures in organisms.
Few groups have the engineering expertise and resources needed to make this kind of artificial intelligence. DeepMind trained AlphaFold 2 on Tensor processing units. Acid sequence training data sets can be proprietary or released under non- commercial licenses.
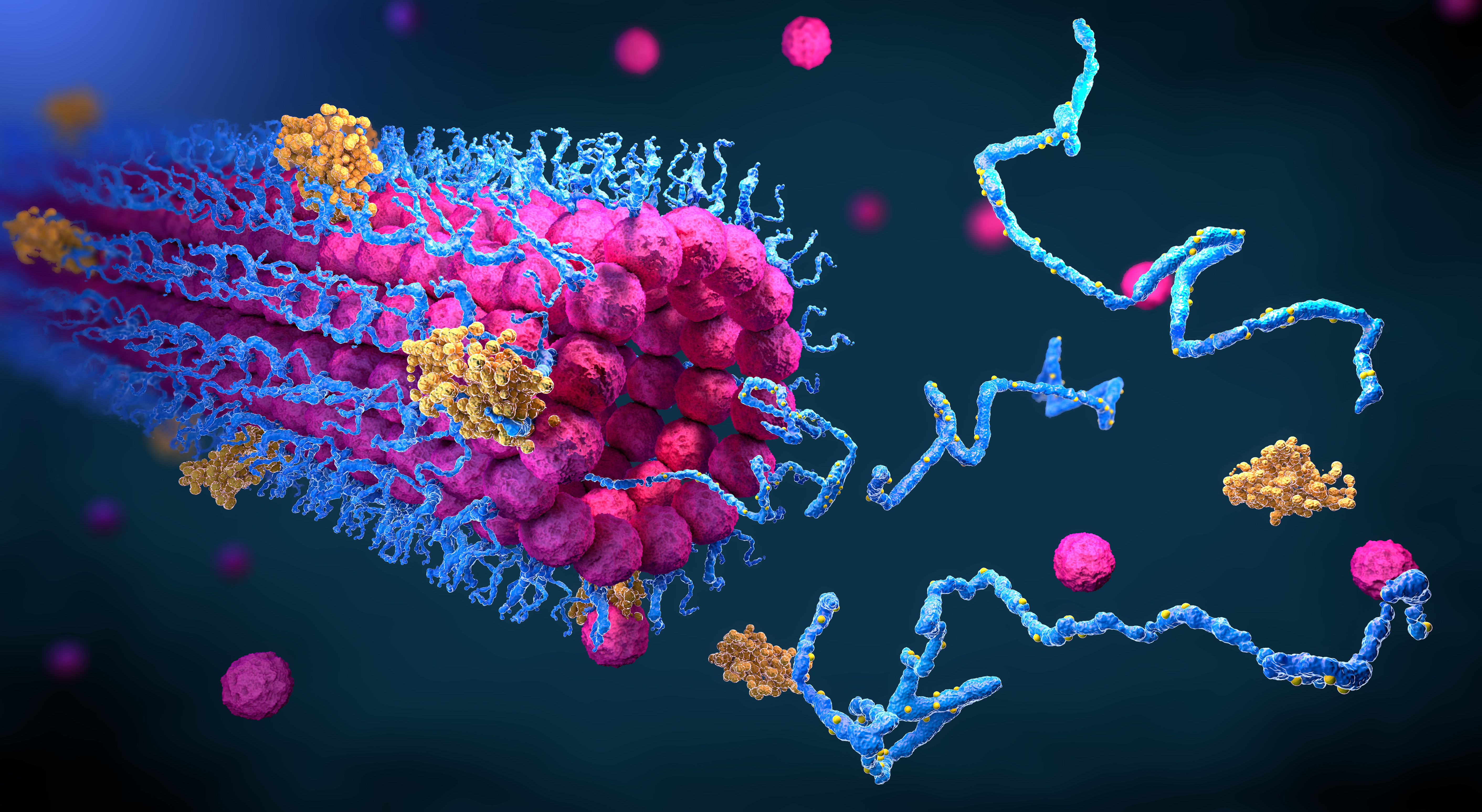
The three-dimensional structure of theProteins The science photo library has image credits.
The community has been able to build on top of the AlphaFold 2 checkpoint released by DeepMind. The model was able to predict quaternary structures, something which few, if anyone, expected the model to be able to do. There are many more examples of this kind, so who knows what the wider scientific community could build if it had the ability to train AlphaFold-like prediction methods.
Two ongoing community efforts to replicate AlphaFold 2 will help facilitate large-scale experiments with various folding systems. A better understanding of what the systems can accomplish and why will be the focus of the researchers.
The project is at its core a project for the community. It could take just one or two months for the first deliverables to be released, or it could take much longer. I'm pretty sure that the former is more likely.
The vaguer mission of the BioLM project is to apply language modeling techniques to biochemical sequence. Several open source text-generating models have been released by Eleutherai, a research group that hopes to train and publish new biochemical language models.
ProGen is an example of the types of work that could be done by BioLM. ProGen distinguishes between words and a sequence of acids. A language model predicting the end of a sentence from the beginning is what the model is trained to predict.
MegaMolBART is a language model that was trained on a database of millions of molecules to find potential drug targets and predict chemical reactions. The company claims that it was able to predict the sequence of more than 600 million genes in just two weeks.
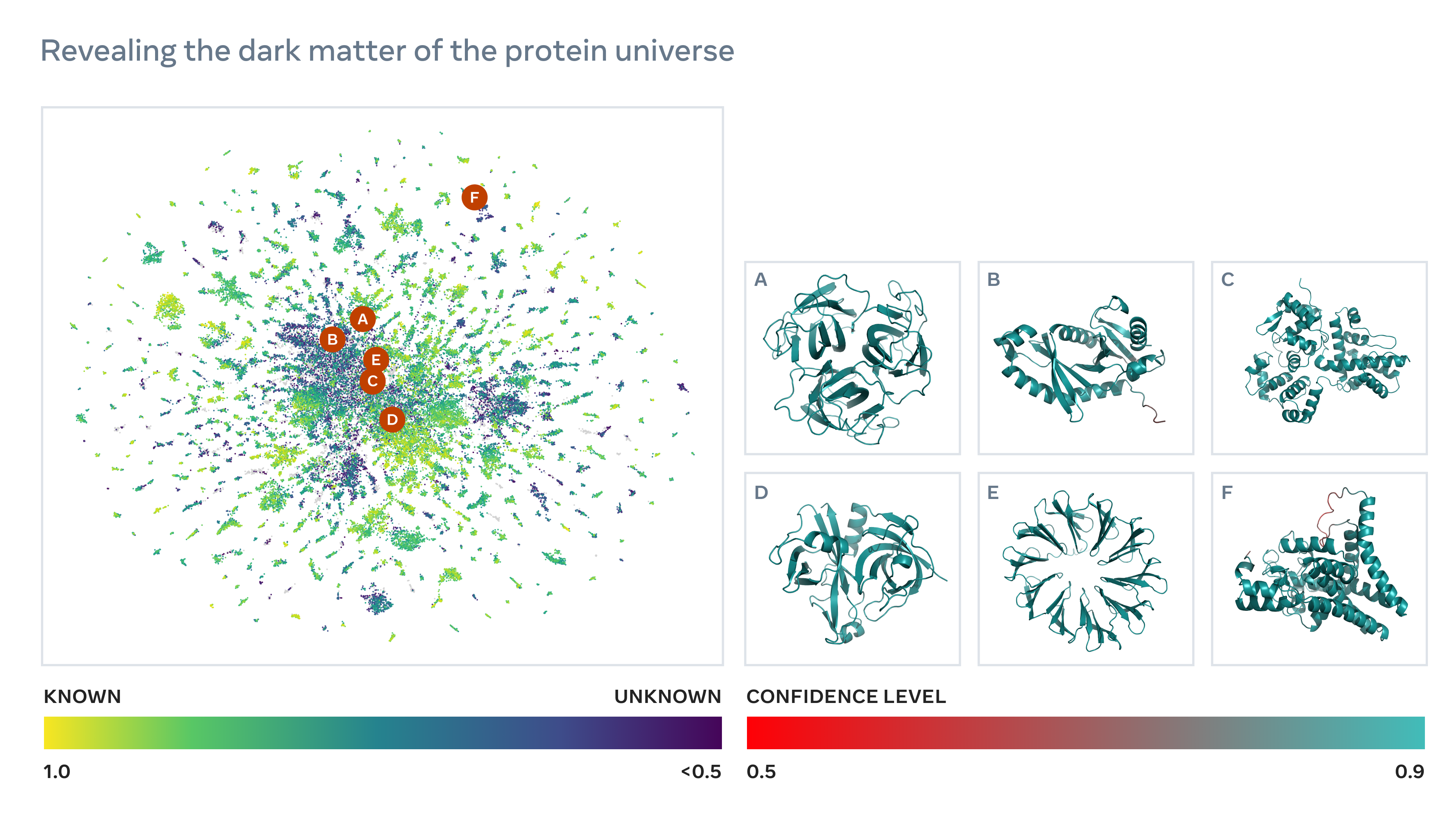
Meta's system predicts the structure of the human body. The image is called Meta.
Mostaque says that they are unified by a desire to maximize the positive potential of machine learning and artificial intelligence in biology and follow in the tradition of open research in science and medicine.
Researchers will be able to gain more control over their experiment for model validation purposes. We want to push the state of the art with more general models, in contrast to the specialized architectures and learning objectives that currently characterizes most of Computational Biology.
As might be expected from a VC-backed startup that recently raised over 100 million dollars, Stability Artificial Intelligence doesn't see OpenbioML as a purely philanthropic effort. When it's advanced enough and safe enough, Mostaque is willing to explore commercializing tech from Open BioML.