It's not possible to move a pharmaceutical scientist from a lab to a kitchen. The same thing happens when it comes to enzymes: They are dependent on the environment. A goal that has remained elusive for over 30 years has been achieved by researchers from Osaka University in a recent study published in the American Chemical Society.
Usually only within a specific cellular environment,enzymes perform impressive functions, enabled by the unique arrangement of their constituents. If you change the environment, the enzyme doesn't function very well. For a long time, a research goal has been to retain or even improve upon the function of the enzymes in different environments. Extensive trial-and-error may have little assurance of achieving an optimal result.
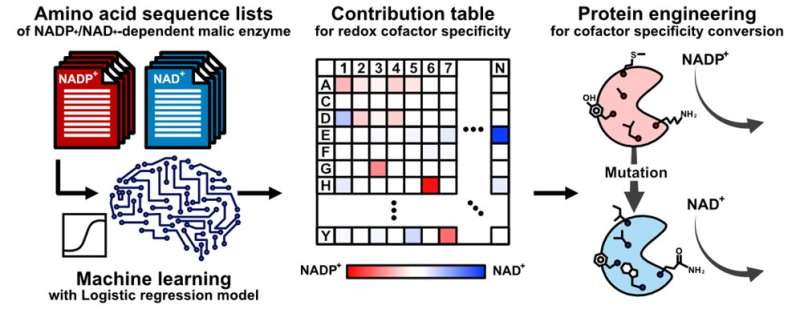
Artificial intelligence can be used to minimize trial and error, but still relies on crystal structures of enzymes which can be unavailable or not useful. Teppei Niide, co-senior author, says that "the pertinent amino acids one should modify in theidase might be best-guesses." To solve the problem, we came up with a method of ranking the amino acids based on the sequence of similar enzymes from other species.
The researchers looked at the specificity of the malic enzyme to the molecule that it transforms and to the substance that helps it happen. The researchers were able to identify the amino acid sequence that did not change over the course of evolution because they didn't change over time.
Hiroshi Shimizu, co-senior author, says that artificial intelligence was used to identify unexpected amino acid residues in malic enzyme. Optimal engineering of the enzyme in laboratories will be made easier by this knowledge.
Artificial intelligence was used to dramatically accelerate and improve the success of substantially reconfiguring an enzyme's specific mode of action. Even in the absence of corresponding enzymes' crystal structures, future advances in engineering will benefit fields such as pharmaceutical and biofuel production.
The Logistic Regression-Guided Identification of Cofactor Specificity-Contributing Residues in Enzyme with Sequence Datasets Partitioned by Catalytic Properties is more information. The book is titled "acssynbio.2c00315."
Journal information: ACS Synthetic Biology