One of the most enduring plant science mysteries, the role of the biological clock, has been solved thanks to a sophisticated computational model developed by scientists.
The effects of a disrupted biological clock can be seen by anyone who has experienced jet lag. Plants suffer when the clock is out of sync.
A permanently jet lag plant is the equivalent of flying from New York to the UK every day. A computer model of the "jetlagged" plant that was able to accurately predict the effect on growth was created by the team.
A feat which has rarely been achieved outside of single-celled microbes is represented by the advance.
The approach, which has been discussed for over a decade, should soon extend to other clock regulated pathways, and lead to fresh insights into wider plant biology that could help improve crop yields and resilience to better cope with climate change.
Plants have a biological clock that is used to detect shifts in the environment and prepare them for changes from dusk to dawn. Although every plant cell has its own clock, which controls 30% of its genes, little is known about their role in plant growth.
Researchers at the University of Edinburgh looked at the effects of clock genes on plants. The clock-mutant plants allowed the team to investigate if clock genes were involved in the release of sugar at night.
Plants need to take care of the energy they capture during the day. Slowing their growth can be caused by releasing sugar too quickly. If the day was 29 hours instead of 24 hours, the clock wouldn't run as fast.
Growth was reduced and night time release of sugars was slower in these Mutants.
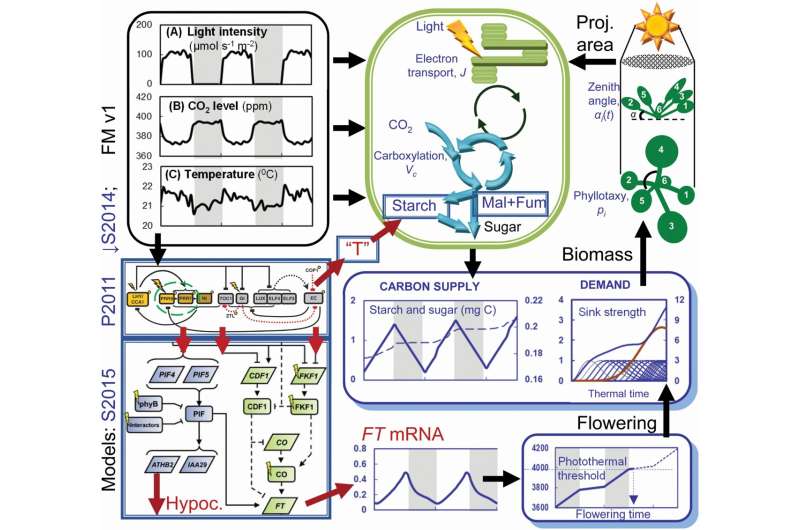
The Framework Model was created by combining mathematical models of clock genes with other models. The Framework Model correctly predicted that the slow release of sugars from starch during the night was responsible for slowing the growth of plants.
Other clock-mutants have been found to interrupt plant growth by disrupting key processes in photosynthesis. The framework model was able to link genes, through measurable pathways, to its impact on the whole plant, a challenge in genetics.
Understanding a human health syndrome caused by a genetic change is similar to this achievement.
The team is going to use the framework model to predict how the plant's genome sequence controls physical characteristics. The approach could lead to a grand unified understanding of biology by showing the interplay between genomes and living systems.
Similar models could be developed to help make sense of the huge amount of data generated by the advances in genome decoding. This type of advance could lead to the unraveling of the complexity of the results in order to understand which are the most important and have the greatest impact on health.
The Framework model shows that we can understand subtle effects at the whole-plant level if we change the timing of genes. We mean to explain and predict. Not all details of this model will transfer to crop species, but it extends the "proofs of principle" for informed crop improvement.
The study was published in a scientific journal.
The Arabidopsis Framework Model version 2 predicts the effects of melatonin on plants. There is a book titled "Insilicoplants/diac010."