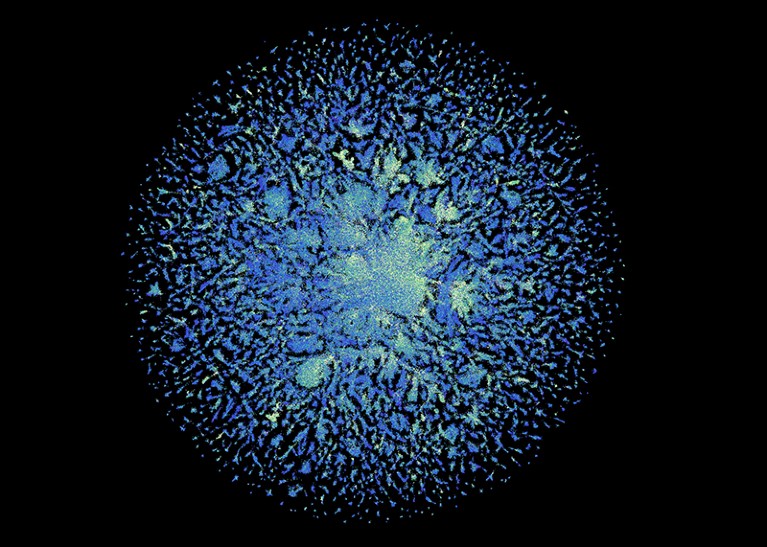
There are structure predictions for 617 million genes. The E SM Metagenomic Atlas is a credit.
Deep Mind revealed predicted structures for 220 million proteins this year, and it covered every single one of them. The dark matter of our universe is being filled in by another tech company.
Artificial intelligence has been used by researchers at Meta to predict the structure of hundreds of millions of organisms.
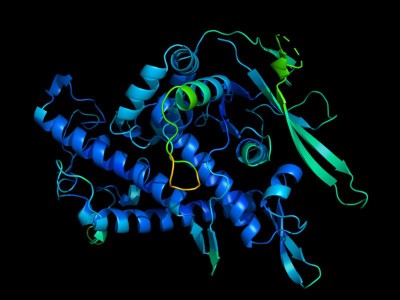
DeepMind's artificial intelligence made a huge leap in solvingProtein structures.
We don't know much about these structures. These are not well known. Alexander Rives is the research lead for Metaai'sProtein team.
The team generated the predictions using a large language model, a type of artificial intelligence that can predict text from a few letters or words.
Language models are trained on a lot of text Rives and his colleagues fed the sequence to the known proteins, which can be expressed by a chain of 20 different amino acids. The network was able to learn to complete the missing parts of the proteins.
The training gave the network an intuitive understanding of the structure of the proteins. The second step is inspired by DeepMind's AlphaFold and combines insights with information about the relationships between known structure and sequence.
AlphaFold is 60 times faster at predicting structures than Meta's network, but it isn't as accurate. Structural prediction can be scaled to larger databases.
As a test case, they decided to use their model on a database of genetic material from environmental sources. Most of the genes come from organisms that have never been cultured and are not known to science.
The structures of more than 617 million genes were predicted. It took 2 weeks for the effort to be completed. The predictions are free for anyone to use.

The next steps for AlphaFold and the revolution of artificial intelligence.
More than one-third of the predictions were deemed to be high quality, which meant that researchers could have confidence in the overall shape of the molecule. In the AlphaFold database of predictions from known organisms, millions of these structures are completely new.
A large part of the AlphaFold database is made of structures that are nearly identical to each other and should cover a large part of the previously unseenProtein universe. More of the darkness can be undone now.
The evolutionary biologist at Harvard University wondered about the hundreds of millions of predictions made by E SMFold. Some might lack a defined structure, at least in isolation. There is still more than half of the space we don't know anything about.
The combination of speed and accuracy of Meta's model was impressed by the Computational Biologist at the Technical University ofMunich. He wonders if it really has an advantage over AlphaFold when it comes to predicting the contents of a metagenomic database. Language model-based prediction methods, including one developed by his team 3, are better suited to determine how a change in the structure of a molecule affects its function. He says that structure prediction will become leaner, simpler and cheaper in the future.
According to a company representative, DeepMind does not currently have plans to include metagenomic structure predictions in its database. Steinegger and his team used a version of AlphaFold to predict the structure of 30 million metagenomic proteins. They are trying to find new forms of their genome-copying enzymes.
Trawling biology's dark matter is the next step for such tools. There will be an explosion in the analysis of these metagenomic structures.