Cornell scientists have created an evolutionary model that connects organisms living in today's oxygen-rich atmosphere to a time when Earth's atmosphere was devoid of oxygen.
Understanding the evolution of these genes will allow us to understand how nature adjusts to environmental changes. Nozomi Ando is an associate professor of chemistry and chemical biology at the College of Arts and Sciences. An analysis of the ribonucleotide reductase family has been published in e Life.
The research school of chemistry at the Australian National University was the first to publish the study. Colin J. Jackson is an author.
It took several high- performance computers a combined seven months to calculate the phylogeny from the 6,779 RNR sequence. The approach is made possible by advances in computing.
RNRs have adapted to changes in the environment over billions of years to conserve their catalytic mechanism. In her lab, she studies how the environment affects the activity of genes. A way to study the relationship between the primary sequence and its three-dimensional structure is provided by the evolutionary information.
According to Ando, RNRs are ideal for finding amolecular records because they have an ancient origin.
"This chemistry would have been required to transition from theRNA world to the DNA world that we are currently living in," Ando said. "Based on the co-factors that RNRs use, it is clear that this family has adapted to the increase in oxygen in the Earth's atmosphere" Billions of years ago, both of these transitions happened.
Scientists calculate how the current sequence came to be when they build a phylogeny of a family of genes. They have to estimate what happened in the past in order to get the sequence that exists today.
Burnim said that the researchers used a dataset of more than 100,000 Sequences and a dataset of 6,779 Sequences to calculate the RNR Family Tree. The sequence lengths range from 400 to 1,100. They compared the sequence with each other to see when they differed.
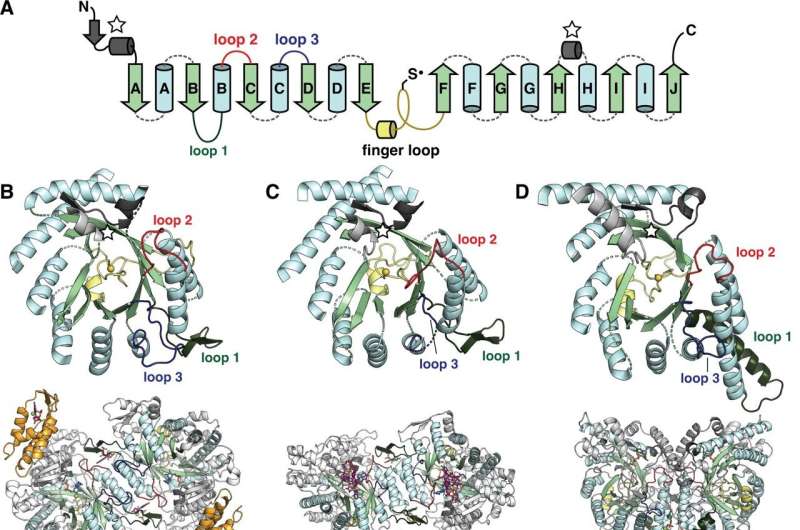
This work led to the discovery of a new group of RNRs that would explain how two different adaptation to oxygen on Earth came about.
The Cornell High Energy Synchrotron Source, the Cornell Center for Materials Research and the artificial intelligence program AlphaFold2 were used to conduct the study.
There was a branch of RNRs that we didn't know was part of the family. Sequences from marine organisms were in this branch. There was an early adaptation to oxygen, according to our characterization of the sequence. The connection between the cyanobacteria and their hosts suggests that they were around at the same time.
The findings support the idea that the adaptation to oxygen took place much earlier than the large-scale environmental changes to the planet.
It is the first unified evolutionary model for all classes of RNRs.
The same approach will be used to study how the same structure evolved to create different reactions.
More information: Audrey A Burnim et al, Comprehensive phylogenetic analysis of the ribonucleotide reductase family reveals an ancestral clade, eLife (2022). DOI: 10.7554/eLife.79790 Journal information: eLife