The institute is part of the Austrian Academy of sciences.
Some of the selfish elements in our genes can jump from one site to another, potentially damaging the genome. Different types of repetitive DNA elements are controlled by the same mechanism in fruit flies.
The effective control of jumping genes is ensured by an uncharacterizedProtein that the researchers named Kipferl. According to the findings, different selfish elements compete for the same part of the genome defense system. The findings can be found in e Life.
A fifth of the fruit fly's genome is composed of genetic parasites that can make copies of themselves and insert themselves into random areas of our genome, potentially disrupting normal genes. These transposons are kept in check by multiple defense mechanisms.
The piRNA pathway is one of the mechanisms that can be used. From sponges to mammals, the piRNA pathway is a smallRNA pathway. The piRNAs are generated from the transposon-rich sequence in the DNA.
The piRNAs are smallRNAs that are used to target the transposons. These piRNAs are used to identify and silence transposons, no matter how far they were able to jump.
The piRNA pathway is found in the fruit fly Drosophila melanogaster. It's not known how Rhino recognizes the piRNA clusters.
The H3K9me3 epigenetic mark has been shown to have an affinity for Rhino in previous studies. This modification is a marker of a type of tightly packed DNA called heterochromatin.
H3K9me3 is found in other parts of the genome, but is not related to piRNA clusters. H3K9me3 is a close relative of Rhino.
It was not clear why HP1 and Rhino bind to different subsets of Heterochromatin even though they both have the same affinity. H3K9me3 was needed, but not enough to explain Rhino's binding to chromatin. There must be more than one cue that helps target Rhino to piRNA clusters.
The team cataloged direct interactors of Rhino in order to find a partner that could guide Rhino to piRNA clusters. The researchers used a combination of genetic, genomic, and neuroimaging approaches to identify Rhino's "companion" in the ovary of the fruit fly.
The Zinc-fingers are used for the sequence specific binding to guanosine-rich DNA. Most piRNA clusters are defined by the combination of the specific DNA binding sites with the local Heterochromatin, according to the team. The interaction between H3K9me3 epigenetic marks and Rhino is stable at these sites.
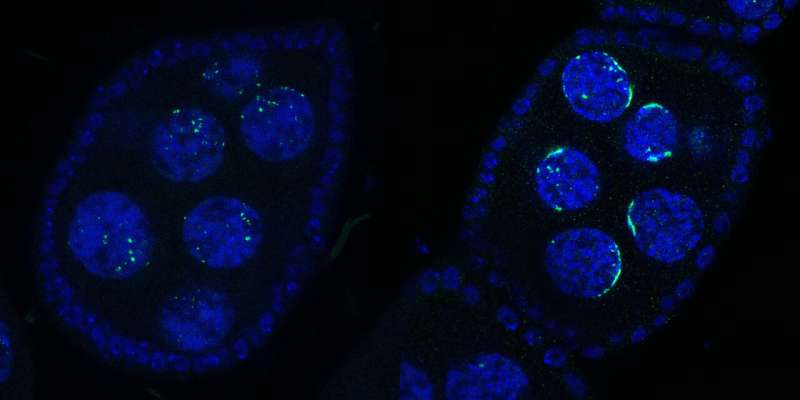
The team knew that Rhino can also be found in piRNA clusters. Rhino was shown to bind to a satellite array. The non-coding and non-transposing genes are located close to the chromosomes.
When we looked at the flies under the microscope, we saw a striking effect on rhino. Rhino was no longer a part of the piRNA clusters when Kipferl was altered. It was accumulated at the satellite array.
This was the first observation that led to the naming of the newProtein'Kipferl'. We found out later that the structures were related to the Satellite array. The scientists showed that Rhinoferl helped to distribute piRNA clusters and avoid its sequestration to the satellite array.
One of the fastest-evolving genes in flies is rhino. The fast evolution may be due to positive evolutionary pressure coming from the satellite array.
The satellite array can recombine." Whole chromosomes could be lost if they do it in an uncontrollable way. The Satellite array might need a control mechanism to package them. This may be the reason why the Satellite array wants to sequester all the rhino.
The piRNA pathways may have different roles in their interactions with the Satellite array. The piRNA pathway is needed to silence the transposons because they pose a danger to the function of the genome.
The piRNA pathway is seen by transposons as the "enemy" that prevents them from spreading. Satellite array simply need an additional layer of control to make sure that they can maintain their high copy number. She says that she would imagine to be a factor that ensured their survival.
The scientists think that the Satellite array might be using another partner to help Rhino get to their genetic material.
We think that it was necessary to help retarget Rhino to the piRNA clusters in order to counter the sequestration of Rhino by the satellite array. Our findings show that Rhino may be caught in a crossfire of genetic conflicts. The fruit fly has both Rhino and Kipferl in it. The first of several Rhino guidance factors may be Kipferl.
More information: Lisa Baumgartner et al, The Drosophila ZAD zinc finger protein Kipferl guides Rhino to piRNA clusters, eLife (2022). DOI: 10.7554/eLife.80067 Journal information: eLife Provided by IMBA- Institute of Molecular Biotechnology of the Austrian Academy of Sciences