The process of translation synthesis is important for survival and every living cell relies on it. One of the most common targets for antibiotics are the machines that translate.
For the first time, scientists led by Julia Mahamid's group at EMBL have visualized at atomic detail how antibiotics affect the process of cell production. It is the first time that scientists have observed atomic-level structural changes in translation machinery directly inside a cell, rather than using isolated molecule in a test tube. They were able to identify mechanisms that machines use to talk to each other.
Researchers from the Max-Planck-Institute for Biophysical Chemistry, Gttingen and the University of Edinburgh were involved in the study. The research was supported by contributions from the Bork and Zimmermann-Kogadeeva groups at the EMBL.
There are minuscule organisms and machines.
Mahamid and her team are experts in studying the bacterium Mycoplasma pneumoniae. Even though it's only ten thousandth of a millimeter in size, the bacterium causes atypical pneumonia in humans.
The first author of the study said that Mycoplasma was chosen because they are among the smallest and most minimal living cells.
A three-dimensional view of a cell can be created by combining serial images of flash-frozen biological samples using an electron microscope. It is possible to capture high-resolution snapshots of different states of a machine in action and combine them into a movie.
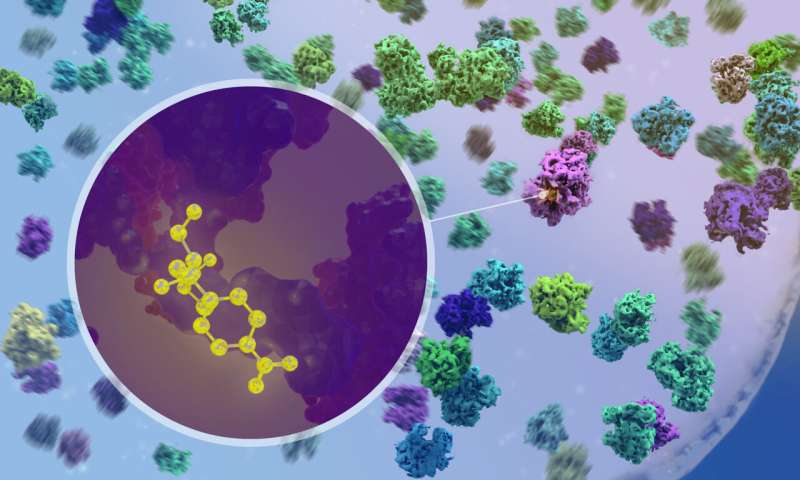
The ribosomes are one of the most prominent structures when looking at a Mycoplasma cell. The ribosome may have existed before the emergence of cells. All cells contain ribosomes, the primary machines involved in the translation of genes.
The Mahamid group's approach allowed them to see the structure of the ribosomes at atomic resolution. The ribosome structure can be deciphered by studying a large number of ribosome frozen at different stages of their activity cycle. They were able to see how the process of translation is organized in the three-dimensional space inside the cell.
"Ribosomes function as highly connected systems, rather than individual machines," said Xue. New features in ribosomes were revealed.
There are antibiotics in action.
The researchers were able to observe what happened when antibiotics entered the cell. They could confirm that the antibiotics chloramphenicol and spectinomycin bind to different sites on the ribosome. It had been predicted by studies on isolated ribosomes, but never before seen in action inside a cell.
When we saw the drug molecule binding to a ribosome, it was exciting. It was exciting when we discovered that the ribosome populations in antibiotic-treated cells are fundamentally reshaped.
The researchers found that the interactions between ribosomes and other complexes in the cell changed in response to the drug. Mahamid said that this can help understand off-target effects of antibiotics and help design combinations of antibiotics to increase their efficiency.
The Mahamid group uses the power of cryo-ET to study important biological processes. Mahamid said that the model system is applicable to many more complex models. In our group, we study the interaction between viruses and their human cell host, the organization of human pluripotent stem cells and the functioning of their ribosomes, and even large multicellular 3D organoids that our collaborators and we grow from cells taken directly from cancer patients.
More information: Julia Mahamid, Visualizing translation dynamics at atomic detail inside a bacterial cell, Nature (2022). DOI: 10.1038/s41586-022-05255-2 . www.nature.com/articles/s41586-022-05255-2 Journal information: Nature