The Receptor on a human cell is one of the key factors that the COVID-19 virus needs to enter a host.
There is no replicating if you don't have a Receptor. There is no infections without replication.
Two years ago, the MacMillan Lab introduced a cellular mapping technology called Map, which has been used by researchers in the Department of Chemistry and the Department of Molecular Biology.
Researchers found that four of them are important.
The research was published in a journal. The tools used to fight the virus could be expanded.
Two years ago, at the height of the Pandemic uncertainty, Alexander Ploss and David MacMillan began the collaborative project.
Since the emergence of the SARS-CoV-1 virus in 2003 scientists have known that its primary entry point was angiotensin-converting enzyme 2. The sameidase was found to be the same as the one that causes COVID-19.
The project assumed that the other stories were not the only ones.
One of the host molecule that the virus depends on to enter into lung cells is called ACE2. Let's see if there's more we can find. We were looking for quick binders.
The entry process is not easy to understand. Along this way it may interact with other host factors, as the virus has to pass through the cell membranes to get into a cell. I don't want to say that everything is decided by the internet. There are a number of processes within the cell that can affect the severity of the disease.
It is the first important step. The game is over if the virus is not able to get in.
Steve Knutson is a co-author on the paper and a research fellow in the MacMillan Lab. Biology can be promiscuous, and we guessed correctly that the SARS-CoV-2 spikeProtein interacts with multiple host cells for entry.
This is a perfect research fit for the Map technology, he said.
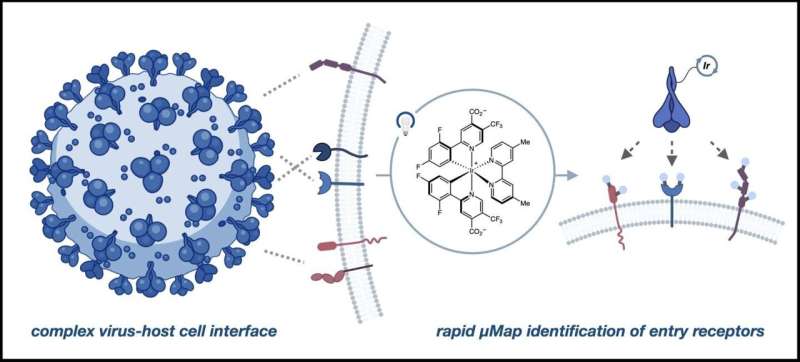
The antenna is called Map.
Micromap is a technology that can be used to identify the neighbors of a cell. A photocatalyst, a molecule that when activated by light spurs a chemical reaction, is used to flag these spatial relationships.
In this work, researchers used the spikeProtein itself as a marker to identify all thereceptor sites in the vicinity ofACE2
Alex had a hunch that there were other things besides ACE2 that could allow him to think about infectious diseases. When we put the photocatalyst on the spike protein, we put an antenna on it to absorb the light.
It can't let that energy go away over long distances. It can't give it to anyone else. The molecule that is free floating has to be very small. We know what's going to happen to it. We are aware of what is happening with it.
Scientists characterized the novelreceptors using a virus pseudoparticle after they were identified. The pseudoparticle does not carry the genetic material to spread the disease. Four entry factors are worthy of further investigation.
The entry process from the infectious cycle can be studied with the help of the pseudoparticle system. If you're looking for the impact of certain host factors on entry, you want to know that you can study it on your own. In this case, we're introducing a reporter gene into the cell and using it to measure how efficiently entry has taken place.
Researchers wonder if there is a clue to disease severity if there is further work done on the function of receptors.
Saori Suzuki is an associate research scholar in the Ploss lab. There were four factors that were outstanding. We need to do more evaluation.
We need to assess how these factors support ACE2 for virus entry and whether newly emerging viral variant use the same set of factors.
More information: Saori Suzuki et al, Photochemical Identification of Auxiliary Severe Acute Respiratory Syndrome Coronavirus 2 Host Entry Factors Using μMap, Journal of the American Chemical Society (2022). DOI: 10.1021/jacs.2c06806 Journal information: Journal of the American Chemical Society