Any sufficiently advanced technology is indistinguishable from magic.
Indika Raja pakse is a believer. A mathematician and engineer is now a Biologist. Blending these disciplines is important to unraveling how cells work.
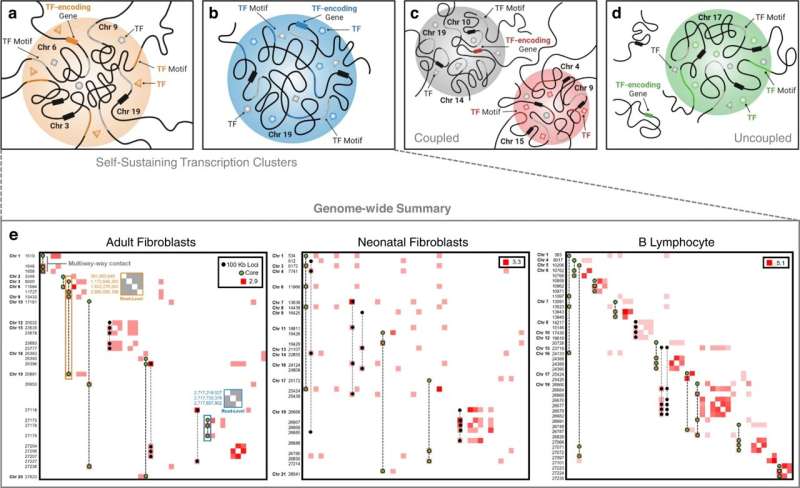
A new mathematical technique is being used to understand how a cell's nucleus is arranged. The self-sustaining transcription clusters, a subset of proteins that play a key role in maintaining cell identity, were revealed by the technique.
They hope that this understanding will expose vulnerabilities that can be used to reprogram a cell.
Cancer biologists think genome organization plays a big part in understanding uncontrollable cell division and whether we can reprogram a cancer cell. Rajapakse said that we need to understand more about what's happening in the nucleus. The U-M has a cancer center.
The paper is in Nature Communications. A group of graduate students led the project.
The team improved upon an older technology to look at which parts of the genome are close together. It's possible to identify those that occur in cancer. It is only able to see the adjacent genomic regions.
The new technology, called Pore-C, uses more data to show how the nucleus of a cell interacts with other parts. Hypergraphs is a mathematical technique used by the researchers. Think of a diagram in three dimensions. It allows researchers to see more than just the pairs of genomic regions that interact.
We can comprehend this multi-dimensional relationship. We can understand organizational principles inside the nucleus with more detail. Rajapakse said that if you understand that, you can understand where the organization's principles deviate. It's like putting three worlds together to study something.
They tested their approach on fibroblasts, adult fibroblasts and B cells. The groups of transcription clusters were identified. They discovered self-sustaining transcription clusters, which are key genes for a cell type.
This is the first step in a larger picture.
To understand how a cell goes through different stages, I want to build a picture over the cell cycle. Rajapakse said that cancer is uncontrollable. We can reprogram systems if we understand how a normal cell changes over time.
More information: Gabrielle A. Dotson et al, Deciphering multi-way interactions in the human genome, Nature Communications (2022). DOI: 10.1038/s41467-022-32980-z Journal information: Nature Communications