A new way hospital-acquired infections resist antibiotics has been found by researchers.
It is possible forbacteria to acquire resistance to antibiotics through random changes in their genes that give them an advantage. To fight back with new drugs, we need to discover how genes helpbacteria survive antibiotic attacks.
The genetic code that leads to antibiotic resistance has been found. They may be present in other infectiousbacteria.
The results of the team led by researchers at Imperial College London were published in the journal.
Resistance is rising.
The researchers looked at the bacterium Klebsiella pneumoniae, which causes infections in the lungs, blood and wounds of those in hospitals, with patients that have compromised immune systems being especially vulnerable.
Carbapenems are a family of drugs used to treat K. pneumoniae. Hospitals use these drugs when other antibiotics fail.
Carbapenem-resistant K. pneumoniae is a critical World Health Organization priority 1 organisms.
In order to be effective, antibiotics need to get inside the bacterium, and in K. pneumoniae this happens via a channel in the outer shell. The team was able to shut down some of these channels and keep carbapenem antibiotics out.
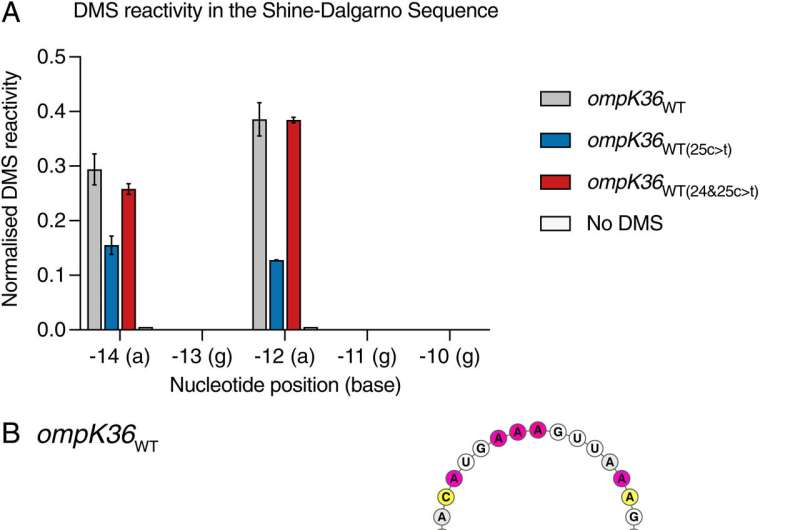
There is a 'Silent'mutation.
The standard mutations that result in resistance are different to this one. When the genetic code is changed so that it is read by ribosomes, it creates a different chain of amino acids.
ribosomes can't read the code and produceProtein from it because of the altered structure of an important mRNA intermediate.
Changes to the amino acid sequence are usually searched for when looking formutations. Since the structure is altered rather than the sequence itself, it could be thought of as a "silent" change.
Dr. Joshua Wong, from the Department of Life Sciences at Imperial, said that in the age of big data andgenomics, the genetic code may be considered silent.
"This discovery should change how we view the genetic code inbacteria and possibly indicates that we in the scientific community have overlooked other similar Mutations that may have important effects." Our work focuses on a single change but fundamentally changes how we interpret others.
The use of antibiotics is driven by this.
The University of Oxford, the University of Florence, and Harvard University collaborated with the team at Imperial to identify the distribution of the mutation and assess resistance levels.
The team used data from resistantbacteria samples collected around the world. The idea that it is driven by the need to defend itself against antibiotics suggests it isn't random.
The novel mechanism is driven by antibiotic consumption and evolved on several occasions independently, according to the leader of the research. The side effects of excessive antibiotic use in hospitals and other settings are highlighted by this.
The team hopes that their finding will be incorporated into bioinformatic tools that analyze genetic sequences to identify the presence of the mutations, as was done with a previous mechanism.
They will continue to work with their partners to find other important changes in the pathogen.
More information: Joshua L. C. Wong et al, Recurrent emergence of Klebsiella pneumoniae carbapenem resistance mediated by an inhibitory ompK36 mRNA secondary structure, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2203593119 Journal information: Proceedings of the National Academy of Sciences