Scientists are using artificial-intelligence tools to come up with new ways to make certain types of cells. Ian C Haydon is a researcher at the Institute forProtein Design at the University of Washington.
South Korean regulators approved the first-ever medicine, a COVID-19 vaccine, in June. The vaccine was created through a labour- intensive trial and error-process that was nearly a decade old.
David Baker, a biochemist at the University of Washington in Seattle, and his team have reported in Science 2 that they can design such molecule in seconds thanks to artificial intelligence.
Artificial intelligence predicts the shape of nearly every knownprotein.
Life scientists are embracing the use of artificial intelligence tools such as DeepMind's AlphaFold. In July, DeepMind revealed that the latest version of AlphaFold had predicted structures for everyProtein known to Science Recent months have seen an explosion in the use of artificial intelligence tools that can quickly dream up new types of drugs. This had been a long and difficult pursuit.
Noelia Ferruz is a computational biologist at the University of Girona, Spain. We are seeing a lot of exciting things.
Most efforts are focused on tools that can help to make original proteins, shaped unlike anything in nature, without focusing on what these molecule can do. A growing number of companies are applying artificial intelligence to the design of proteins to find ways to clean up toxic waste and treat diseases DeepMind in London is one of the companies that is working towards this goal.
The methods are very effective. They are going to become more powerful. What problems are you going to deal with?
Over the past 30 years, Baker's lab has made new proteins. The software he started developing in the 1990s splits the process into several steps. Initially, researchers conceived a shape for a novelProtein, often by cobbling together bits of other proteins, and the software deduced a sequence of Amino Acids that correspond to this shape.
The first draft of the proteins were stuck in different confirmations when they were made in the lab. It was necessary to change the sequence so that it folded into a single structure. An evolutionary biologist at Harvard University who used to work in Baker's lab said that the step involved simulating all the ways in which different sequence might fold. Ten thousand computers would run for weeks doing this.

The next steps for AlphaFold and the revolution of artificial intelligence.
The time consuming step has become instantaneous thanks to the tweaking of AlphaFold and other programmes. One approach developed by Baker's team is called hallucination, which alters the structure so that it becomes ever-moreprotein-like, as judged by the network's predictions. Baker's team created more than 100 small, hallucinated proteins in the lab and found some that resembled the predicted shape.
AlphaFold and RoseTTAFold were trained to predict the structure of individual proteins chains. Researchers found that the networks could model the assembly of multiple interacting proteins. Baker and his team were confident that they could hallucinate multiple copies of a single molecule into different shapes and sizes, just like the COVID-19 vaccine does.
The source is from N. Ferruz et al.
None of the 150 designs the microorganisms were told to use worked. The gunk at the bottom of the test tube was the reason they didn't fold.
The inverse folding problem is a problem that machine-learning scientist Justas Dauparas is trying to solve. While maintaining the molecule's overall shape, the network can act as a'spellcheck' for designerProteins created using AlphaFold and other tools.
When Baker and his team applied the second network to their hallucinated nanoparticles they were able to make the molecule. The researchers used a variety of experimental techniques to determine the structure of 30 of their new proteins. Giant rings with complex symmetries are not found in nature. A biophysicist who co-led the effort says that the approach could be used to design nanoparticles that correspond to almost any shape. The networks can do a lot.
Elofsson says that deep-learning tools have been a game-changer in the field. Push a button and you get something that works one in 10 times. Baker's team did a great job of combining multiple neural networks to tackle different parts of the design process. We now have complete control over the shape of theProtein
Baker's isn't the only lab that uses artificial intelligence. More than 40 artificial intelligence tools have been developed in the last few years, according to a review paper posted to the bioRxiv this month.
The inverse folding problem is tackled by many of these tools, using approaches borrowed from image-recognition tools. The architecture of some of them is similar to that of a language neural network such as GPT-3, which can produce human-like text. Ferruz has co-developed one of these networks.
A machine-learning researcher at the University of California, Berkeley, who developed an inverse folding network with researchers from Meta 7, says that it's not always clear how to compare tools.
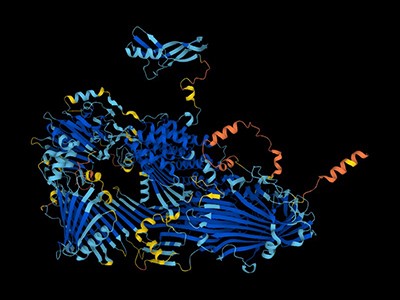
There are four examples of the phenomenon ofhallucination. AlphaFold changes the sequence until the software confidently predicts that it will fold into a well-defined 3D shape. Predictability is shown by the colors red, yellow, light blue, and dark blue. The frames have been slowed down to make them clearer. The credit is given to Sergey Ovchinnikov.
Many teams look at their network's ability to determine the sequence of an existingProtein from its structure This doesn't apply for all methods and it's not clear how this metric applies to the design of novel proteins. Ferruz would like to see a competition similar to the Critical Assessment ofProtein Structure Prediction (CASP) experiment in which AlphaFold first demonstrated its superiority over other networks. I think it is a dream. She says that a CASP would move the field.
The ultimate test of Baker and his colleagues' methods is the creation of a novelProtein in the lab. This is shown by their initial failure to make a hallucinated assembly. BasileWicky, a biophysicist in Baker's lab who co-led the effort, says thatAlphaFold didn't work in the wet lab.
Jinbo Xu, a Computational Biologist at the Toyota Technological Institute of Chicago in Illinois, said that not all scientists have easy access to experimental set ups for their work. Finding a lab to work with can take a long time, which is why Xu is establishing his own wet lab.
When it comes to designing proteins with specific tasks in mind, experiments will be important. In July, his team described a pair of artificial intelligence methods that allow researchers to put a specific sequence or structure in a novelProtein 8 A vaccine against a respiratory virus that is a leading cause of infant hospitalizations is one of the things that could be done with these approaches.
Isomorphic Labs in London was launched last year by DeepMind, which plans to use artificial intelligence to find drugs. According to DeepMind's chief executive, Demis Hassabis, there is an obvious and promising application for deep- learning technology, and specifically for AlphaFold. There is a lot of work being done in the space. It's early days.