Plants are able to reprogram their genetic material to survive and thrive when faced with environmental challenges such as dry weather. The chemical code that causes those changes can be deciphered and duplicated to breed more crops.
That's the conclusion of a team of Penn State geneticists that conducted the first-ever study of the effects of reprogrammable plants. Understanding and eventually exploiting that process will be important to breeding crops that can tolerate weather extremes brought on by climate change.
Sally Mackenzie is a professor of plant science in the College of Agricultural Sciences and a professor of biology in the Eberly College of Science. We don't have to cross breed to get it done. Plants go into that state on their own, so we don't need to add new genes.
The ability to adapt relatively quickly to environmental change through these stress-induced states is passed down through many generations.
Previously, the relationship between epigenetic changes and gene networks that influence plant growth had not been studied. She said that this type of research takes specialized methods of data analysis and massive computational capacity at Penn State.
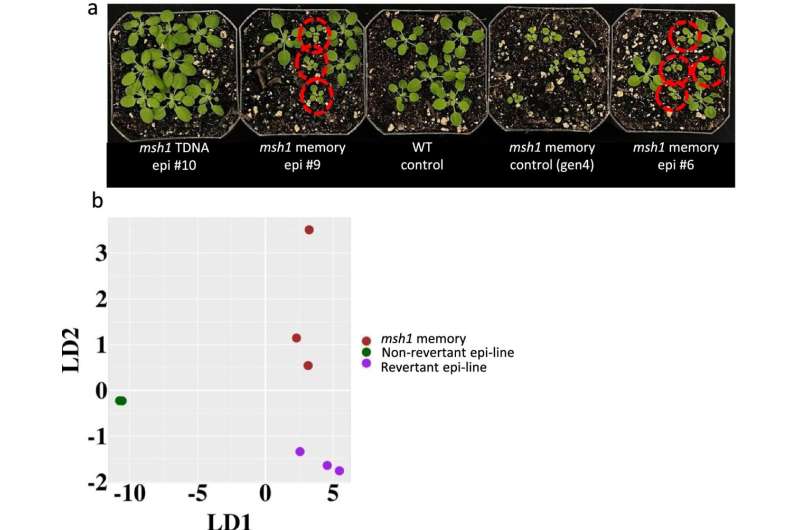
The research group discovered that they could control a broad array of plant-resiliency networks by manipulating a single gene. After the MSH1 gene is silenced, the plant can adjust its growth, change root configuration, limit above-ground biomass, and alter its response to environmental stimuli.
The potential for this technology is much bigger than plant science.
She said that this is not just a feature of the MSH1 system. Our study likely will allow us to use this newfound decoding method to help in early disease diagnostics. The methylome will provide an indication when a biological system is malfunctioning or modified.
The study was led by Hardik Kundariya, a senior researcher in the Eberly College of Science and a member of the group. The relative of cabbage and mustard in the Brassica family is one of the model organisms used for studying plant biology.
The MSH1 gene was manipulated by the researchers to cause at least four different nongenetic states to impact plant stress response. They were able to locate and decode data relevant to plant growth from the cross-comparing data.
The researchers reported that manipulating that gene resulted in changes in the plant's appearance and growth characteristics that could help it survive.
More information: Hardik Kundariya et al, Methylome decoding of RdDM-mediated reprogramming effects in the Arabidopsis MSH1 system, Genome Biology (2022). DOI: 10.1186/s13059-022-02731-w Journal information: Genome Biology