Structural shifts that offer clues about how the COVID-19 coronaviruses works were discovered by scientists. The findings may inspire the design of new drugs to counteract COVID-19 and help head off future coronaviruses.
The City University of New York and the U.S. Department of Energy collaborated to conduct a study on the coronaviruses at body temperature.
Most of the structures we have seen come from frozen samples. You should get a more realistic picture of what's happening during an actual infection if you're working at a temperature like that.
He said that the team used temperature as a tool. We can learn a lot about the mechanics of theProtein by turning that knob.
Figuring out Mpro's structure.
The main protease of the viruses that cause COVID-19 is the Mpro. It's a protease that cuts other proteins. In many viral infections, the cells that have been bitten produce a virus's functionalProtein as one single connected chain. The pieces of the virus are cut apart by the proteases. COVID-19 could be put on hold if a drug is found to disabling Mpro.
X-ray crystallography is a technique used to study the structure of an object. The Office of Science uses the facility to produce x-rays. It is possible to see the three-dimensional arrangement of atoms on a sample of a biological molecule.
It can be difficult to study samples that are not frozen.
The higher the temperature, the greater the chance of x-rays damaging the crystal.
The crystal is moved linearly as it moves through the x-rays. The X-ray dose is distributed over the entire length of the crystal.
The small size of the X-ray beam makes it possible to focus the beam on the smallest part of the crystal as it rotates.
One full dataset can be collected in just 10 seconds per sample, with good quality to solve a structure before significant X-ray damage occurs.
From cold to warm.
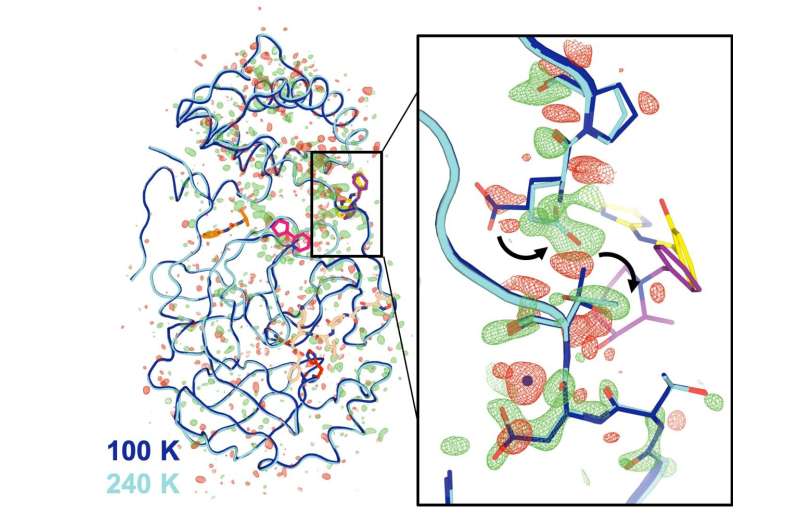
The first-ever series of Mpro crystallographic data was collected at five different temperatures, ranging from -28 degrees F to 39 degrees F. Under high humidity, they studied the room- temperature crystal. They fed the data into a computer simulation that identified many possible atomic-level arrangements.
The results showed a change in the structure of the molecule. Some of the features that the team saw were unique to the enzymes.
Most of the changes were outside of the active site. They were far away from that location. The data suggests that these distant sites could affect the active site through a type of remote-control mechanism that is common in biological systems. The function of the enzyme could be blocked by disabling even those distant locations.
Mpro is a kind of folded ribbon, made up of two identical halves that bind together in a symmetrical way. The "dimer interface" links to the active site through a flexible loop region of theProtein.
The scientists found that the grip of the handshake changes at higher temperatures. There may be some kind of communication between the active site and the dimer interface when the virus is present.
Drug design has a path to it.
Drug design is dependent on subtle changes that are less than a billionth of a meter.
Other studies have shown that small drug-like molecule can bind to the enzyme at a number of different locations.
We could potentially have a new foothold for altering the function of the enzyme if we could perfect these molecules, elaborate on them, and tinker with them. Our findings inspired us to explore this idea.
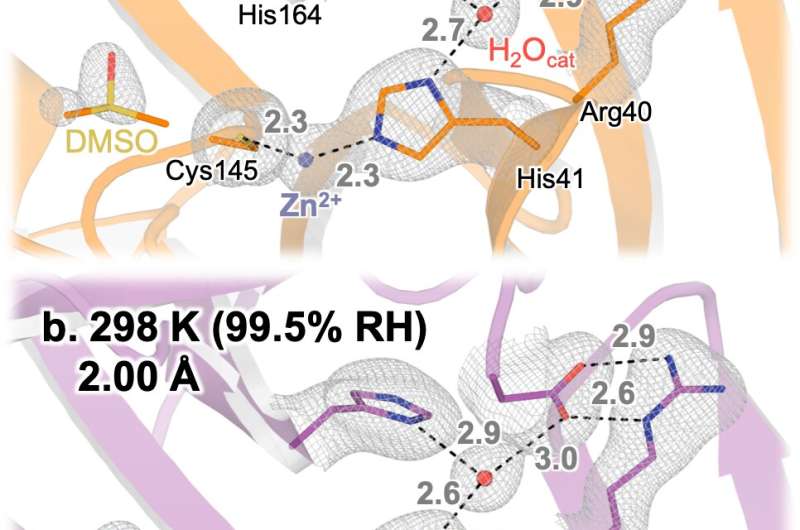
Water has a role to play.
It's possible to explore the enzyme at elevated humidity and see how it behaves in water-filled cells.
After selecting the crystal we want to study, we put a special sleeve over it to keep it from drying out. After putting the sample in the beamline for X-ray data collection, we remove the sleeve and blow a continuous stream of air over the crystal as we collect the data.
Under high-humidity conditions, the water molecule that bind to the enzyme was found.
Andi said that the water molecule give you a hint.
He and his group have been studying a range of potential drug-like molecules that seem to replace weakly bound water molecule when they bind near Mpro's active site. A paper about that work was recently published in a journal.
Both scientists were grateful for the collaborative spirit of the entire team, which included additional scientists at both Brookhaven Lab and CUNY.
"Remote collaboration was very important to make this project work, where we had people on site at the beamline, and people offsite who could do the computer modeling." There are many examples of the scientific community coming together.
More information: Ali Ebrahim et al, The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (Mpro), IUCrJ (2022). DOI: 10.1107/S2052252522007497 Journal information: Scientific Reports