A team led by UT Southwestern researchers has found out how the virus that causes COVID-19 builds a structure that is critical for successful viral replication. It could lead to new strategies to attack COVID-19, which has killed more than 6 million people around the world so far.
A new approach to treat COVID-19 could be offered by exploiting and making drugs against this protein domain.
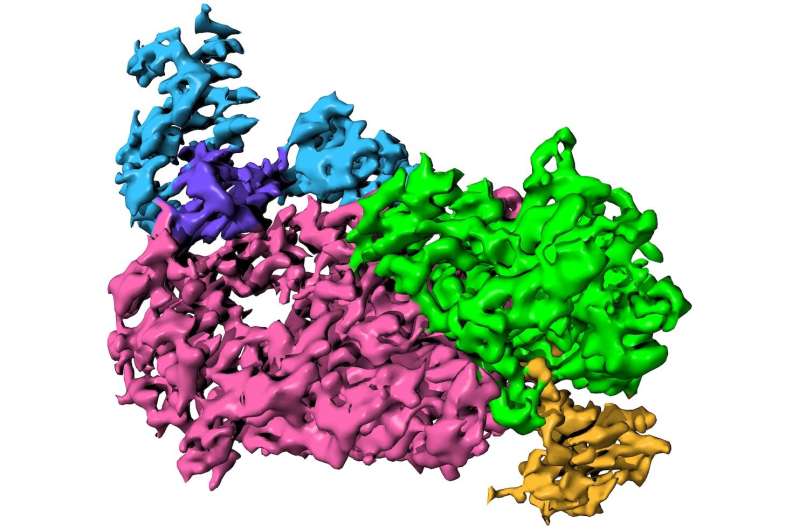
The genetic molecule ribonucleic acid is used to give instructions to the host cells. Dr. Tagliabracci explained that a cap on one end of the virus serves multiple functions. It hides theRNA from the host cell's immune system, protects it from attack by exonucleases, and recruits cellular factors that use theRNA to make viral proteins He said that if the virus is missing the processes will stop and the infections will stop.
The new study suggests that the nsp12 is involved in the synthesis of the RNA cap. The main focus of the Tagliabracci lab is the NiRAN domain. Experiments show that the NiRAN domain transfers the viral RNA to the nsp9 to create a transcript that is crucial for cap formation.
According to Dr. Tagliabracci, he and his colleagues are looking at ways to stymies the NiRAN domain function, which could lead to a new class of drugs.
He said that the long-term fight against COVID would need antivirals targeting different parts of the viral life cycle. Adding a capping inhibitor would be great.
More information: Gina J. Park et al, The mechanism of RNA capping by SARS-CoV-2, Nature (2022). DOI: 10.1038/s41586-022-05185-z Journal information: Nature