The atomic structure of proteins has been probed to add to evidence that the wobbles, shakes and quivers of the molecule play a critical role in their function. Scientists may be able to design new drugs that can modify or disrupt the intricate "dances" of proteins to alter their functions, thanks to the findings of the research.
The results of the researchers' work will be published in July.
Structural components of tissues, along with the "business ends" of biology, are made up of organic compounds with blueprints in DNA.
Scientists have debated the significance of this "dancing" act, though it has been known for a long time. "Protein wiggles are critical for this communication, and the way they engage with the right partner at the right time is very important for understanding their function," he said.
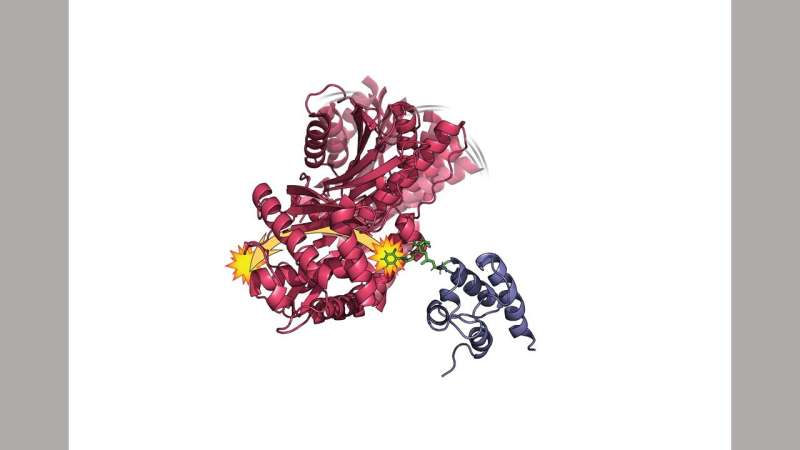
Frueh's team studied the wiggling action of the HMWP2protein, which is a type of non-ribosomalpeptide synthesizer. Complex natural products from small chemicals can be made with the help of these enzymes.
Natural products can have pharmaceutical properties. Yersiniabactin is a molecule that is used in the treatment of urinary tract infections and bubonic plague.
Scientists can modify the domain to make them produce new chemicals if they understand how it works.
The team found that the widespread wiggling of one domain leads to a process that allows the domain to connect with several partner domains at the same time.
Nuclear magnetic resonance (NMR) spectroscopy, a device that uses powerful magnetic fields to probe themolecular environments of nuclei within the center of atoms, was used to track the motion of one of the domain's down to each individual atom.
Tracking the motions of large proteins with the device is difficult because of its small size. Frueh's team used nitrogen-15 and carbon-13 to overcome the challenge.
When the second domain is not modified, the two domains will only bind to one another, so they won't waste time together. The first domain is able to sense when the second domain is changed and we wanted to know if motions played a role in this recognition process.
There were changes in motion across the entire domain, not only where it binding to the second domain but also at a second binding site used by a third domain.
About 40 billionths of a meter is how far apart these two sites are. The scientists were interested in how they interact.
The sites' ability to interact with other domains weren't blocked by the Mutation.
Frueh says that all of the movement of theProtein domain was hampered. According to the researchers, the lack of movement within the protein made it hard for it to bind with other domains.
Frueh points out that scientists could design new drugs that don't target the natural active site but instead stop the movement of the molecule to inactivate it, if they knew more about the movement of the molecule. He says that such an approach could allow for more flexibility in the design of drugs.
Frueh says that researchers are studying how computation and artificial intelligence can improve the understanding and prediction ofProtein movement
More information: Subrata H. Mishra et al, Global protein dynamics as communication sensors in peptide synthetase domains, Science Advances (2022). DOI: 10.1126/sciadv.abn6549. www.science.org/doi/10.1126/sciadv.abn6549 Journal information: Science Advances