Cells are able to respond to their environment with the help of cell surface binding agents. MIT chemists have discovered how one of thesereceptors changes its shape when it binding to its target, and how those changes cause cells to grow and flourish.
The target of several cancer drugs is the EGFR, which is overexpressed in a lot of cancer. Tumors can become resistant to these drugs at first. An associate professor of chemistry at MIT says that understanding the mechanism of thesereceptors may help designers of drugs that can evade resistance.
She says that thinking about more general mechanisms to target EGFR is an exciting new direction.
The senior authors of the study are Schlau-Cohen and BinZhang. The paper is written by a graduate student and a former MIT employee.
There is a shape-changingreceptor.
Control of cell growth is aided by the EGFreceptor. It can respond to a variety of growth factors in addition to EGF. Lung cancer and glioblastoma are two types of cancer that overexpress the EGF receptor.
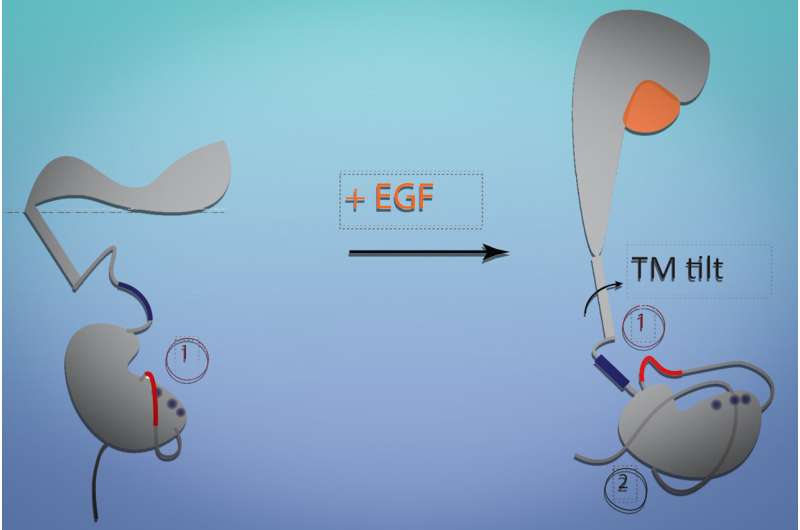
The cell is covered by the EGFR. An extracellular region of the receptor interacts with its target molecule, a transmembrane section is embedded within the membranes, and an intracellular section interacts with cellular machinery that controls growth pathways
The transmembrane and intracellular sections can't be studied because they are more disorganized.
Five years ago, Schlau-Cohen tried to learn more about those structures. A special type of self-assembling membranes called a nanodisc mimics the cell's outer shell. She used a single molecule FRET to study how thereceptor's structure changes when it is bound to EGF.
FRET is used to measure distances. Under a variety of circumstances, the researchers were able to measure the distance between the end of the intracellular tail and the cell's surface.
The researchers were surprised to find that the binding of EGF led to a change in the structure of the receptor. Most models of signaling involve interaction of multiple transmembrane helices to bring about large-scale changes, but the EGFreceptor, which has only a single helical segment within the membrane, appears to undergo such a change without interacting with otherreceptor molecules.
It was surprising to us that a single alpha helix was capable of transducing such a large rearrangement.
Modeling is done in the form of a molecule.
To learn more about how this shape change would affect thereceptor's function, Schlau-Cohen's lab collaborated withZhang's lab. Modeling how a system changes over time can be done with this type of modeling.
The model shows that when the receptor is not bound, it lies flat against the cell. Similar to a hinge closing, when the receptor falls flat, it tilts the transmembrane segment and pulls the intracellular segment close to it. The machinery needed to launch cell growth is blocked by this. Growth signaling pathways are activated when EGF binding is present.
The model used by the researchers shows that positively charged amino acids in the intracellular segment are key to these interactions. The researchers switched the amino acids from charged to neutral.
The consistency between the simulation and experiment is nice. We can use the simulations to figure out what the essential and non-essential elements are for thecoupling. The predictions turned out to be correct, according to the lady.
Cetuximab, a drug that binding to the EGF receptor, prevents this change from happening. Cetuximab can be used to treat colorectal or head and neck cancer, but it can also be used to treat tumors that are resistant to it. Researchers say that learning more about the mechanism of how EGFR responds to different ligands could help them design drugs that are less likely to cause resistance.
More information: Nature Communications (2022). DOI: 10.5281/zenodo.6564353 Journal information: Nature Communications