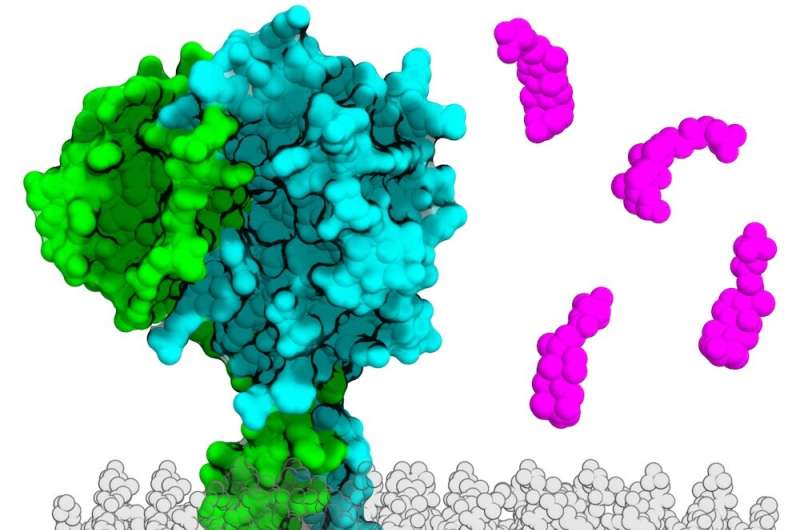
UT Southwestern researchers used artificial intelligence to discover a new family of genes in entericbacteria that are linked by structure but not genetic sequence. A new way of identifying the role of genes in unrelated species is offered by the findings.
In reverse of how it's usually done, we identified similarities in these genes. Instead of using sequence, Lisa looked for matches in their structure.
Dr. Orth's lab studies how marine and estuarybacteria cause infections. The structure of the VtrA and VtrC complex was characterized in 2016 by Dr. Orth and her team. She and her team found the VtrA/VtrC complex in V. parahaemolyticus, which causes food poisoning from contaminated shellfish.
Although VtrA and ToxR share some structural features, it's not clear if a homolog for VtrC also exists in this or any other bacterium.
Dr. Kinch said they had never seen VtrC. We thought that it must exist.
The AlphaFold software was released two years ago and was used to find genes with sequence identities similar to VtrC. This artificial intelligence program is able to accurately predict the structure of some proteins based on the genetic sequence that codes for them.
Even though VtrC and ToxS do not have the same genetic sequence, they are both similar in structure and function. The researchers were able to find VtrC in several other entericbacteria that cause human disease, including the bubonic plague. The roles of the VtrC homologs could be the same as those in V. parahaemolyticus.
Structural similarities could lead to the development of pharmaceuticals that treat conditions caused by different infectious organisms.
More information: Lisa N. Kinch et al, Co-component signal transduction systems: Fast-evolving virulence regulation cassettes discovered in enteric bacteria, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2203176119 Journal information: Proceedings of the National Academy of Sciences