Scientists have been able to figure out the human nuclear pore complex's architecture in greater detail thanks to the combination of AlphaFold2 and experimental and Computational techniques.
The nucleus of the human is separated from the rest of the body through the use of a nuclear pore complex. It is a gateway and a checkpoint formolecules that travel between the nucleus and the cytoplasm. Gene expression and translation are important processes in the cell. The nuclear transport system is involved in a number of diseases.
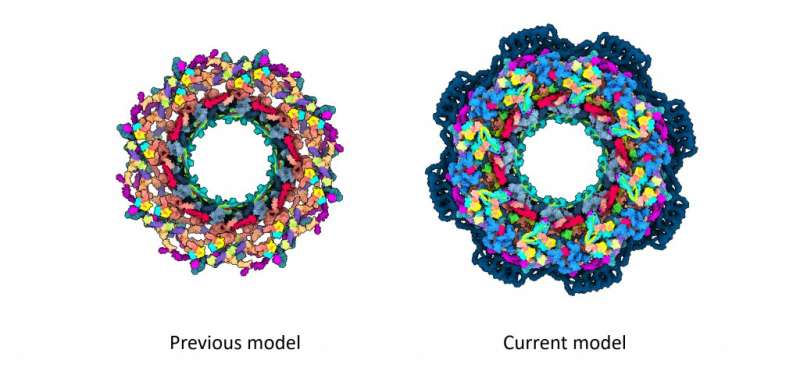
The structure of the NPC is not known. How are its cells made? How does it connect to the nuclear device? These and other questions have been answered by the Kosinski Group at the Centre for Structural Systems Biology. In order to create the most complete model of the human NPC, they combined the AlphaFold2 program with techniques such as single particle cryo-EM.
Structural biologists like the human NPC because it is a 3D puzzle with around 30 differentproteins in multiple copies. A round core is formed with around 1000 puzzle pieces. The most accurate models only covered a small portion of the structure. Scientists have been working on a new model of the NPC structure for two decades.
The new model eliminates much of the ambiguity that existed when the previous models were proposed.
When you disassemble and reassemble an electronic device, it's similar. Jan Kosinski, who co-leads the investigation, said that there will always be some screws left. We know where they are, what they do, and how after we fit most of them.
Artificial intelligence works with experimentation.
The scientists achieved this. It was important to combine several methods. Scientists were able to see the NPC at different levels.
The researchers used a method called cryo-electron tomography to model the NPC. They were able to see the NPC in its cellular environment. AlphaFold2, an artificial intelligence program created by DeepMind, revealed more information about the individual building blocks.
"AlphaFold2 was a breakthrough moment for us," said Agnieszka Obarska-Kosiska, a PhD student. We didn't know the structure of a lot of the genes. You can't put together a puzzle if you don't know what the pieces are. We were able to predict those shapes thanks to AlphaFold2.
The researchers used a version of AlphaFold2 modified by the scientific community to model interactions. They were able to see how the different puzzle pieces combine to form smaller subcomplexes and how those subcomplexes are then glue together to form theNPC.
The Kosinski Group's software Assembline was used to put all the pieces together.
The model was so complete and detailed that the researchers were able to create time-resolved simulations that explain how the nuclear membrane interacts with the NPC, and how it responds to mechanical stimuli.
The future directions.
The work was a big step forward for the research.
Structural biology will embrace cell biology to create atomic models of ever larger assemblies of molecules that perform different functions in different parts of the cell according to Martin Beck. It is now possible to build a complete dynamic model of the NPC and simulation of nuclear transport in atomic detail.
Automatic methods for integrating structural and microscopy data using AlphaFold2 will be developed by the Kosinski Group. They plan to use these approaches to study the causes of viral infections.
The research was published in a scientific journal.
More information: Shyamal Mosalaganti et al, AI-based structure prediction empowers integrative structural analysis of human nuclear pores, Science (2022). DOI: 10.1126/science.abm9506 Journal information: Science