Researchers at the John Innes Centre have developed technology that shows the impact of environmental conditions on the structure of living cells.
The research is the result of a collaboration between two groups of people. This knowledge could be used to fine- tune crops or develop therapies for diseases such as COVID-19.
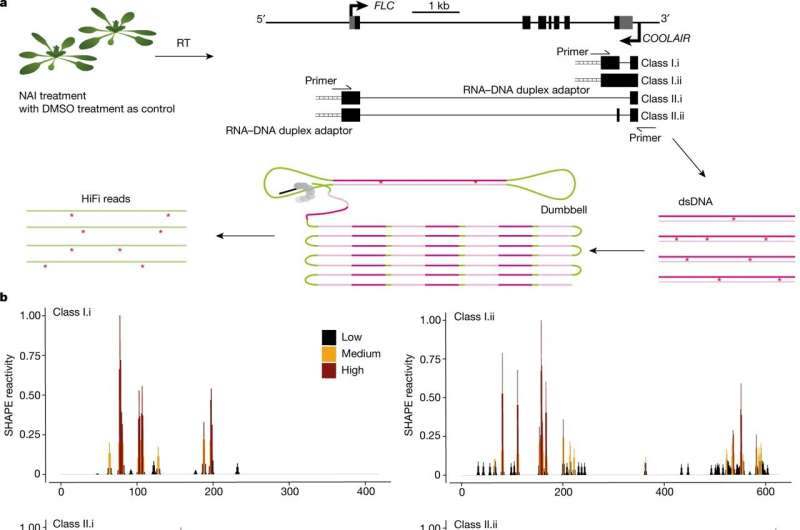
Two important genetic elements COOLAIR and FLC play a role in regulating plant responses to warm and cold. It was not clear how the structure of COOLAIR contributed to the regulation of FLC.
A new technology developed by researchers in the Ding group is able to identify the structure of a single molecule in live cells.
They were able to observe structural changes using this method. The shapes and proportions of the plants change after they are exposed to cold temperatures.
They noticed that the expression of FLC changed when there was a change in theRNA conformations. The researchers were able to change the flowering time of the plants by changing the sequence of theRNA region.
Dr. Ding says that their work shows thatRNAs can adopt different structures. In response to external conditions, these different conformations change. The flowering time of the plant was changed by tuning theRNA structure.
The ability to engineer plant genomes at theRNA cellular level increases the possibility of designing crop types with more desirable agronomic and nutrition characteristics.
The group says that the technology can be used to apply it to human cells.
According to the first author, everyRNA is likely to have its own structure and diversities. The technology we have will allow us to look at the functional importance of the RNA structures.
The group wants to share their technology with other people.
During the process of expression of genes, the genes are transcribed into a form ofRNA. Since it is a single stranded molecule, it is often referred to as the "skinny molecule", but recent work has highlighted its structural diversity and how it affects genes.
When a plant reaches a required level of cold exposure, FLC acts as a brake on flowering, a key part of a molecular mechanism. COOLAIR is antisense to FLC and prevents it from being transcribed after cold exposure. Knowledge of these mechanisms will help understand the effects of climate change.
Nature contains the research.
More information: Caroline Dean et al, In vivo single-molecule analysis reveals COOLAIR RNA structural diversity, Nature (2022). DOI: 10.1038/s41586-022-05135-9. www.nature.com/articles/s41586-022-05135-9 Journal information: Nature